library(clusterProfiler)
library(enrichplot)
library(pathview)
library(org.Hs.eg.db)
library(ggplot2)
library(ggrepel)
library(msigdbr)
library(tidyverse) # for bonus code/dplyr/pipe
# set seed
set.seed(1234)SIB - Enrichment Analysis
Exercise 1
# Import DE table:
NK_vs_Th <- read.csv("data/NK_vs_Th_diff_gene_exercise_1.csv",
header = T
)
# Look at the structure of the data.frame:
head(NK_vs_Th) ensembl_gene_id symbol logFC t P.Value p.adj
1 ENSG00000000003 TSPAN6 -5.6436044 -4.672128 0.0000426000 7.358019e-04
2 ENSG00000000419 DPM1 -0.1818981 -1.101831 0.2780198240 5.176076e-01
3 ENSG00000000457 SCYL3 0.4969874 1.491035 0.1448690710 3.449889e-01
4 ENSG00000000460 C1orf112 1.1217991 1.445899 0.1570598770 3.630935e-01
5 ENSG00000000938 FGR 10.6706873 7.212342 0.0000000198 1.718657e-06
6 ENSG00000000971 CFH -3.4129277 -2.788887 0.0084803000 4.610083e-02# Search for a gene symbol in the data.frame, eg NCAM1 (CD56)
NK_vs_Th[which(NK_vs_Th$symbol == "NCAM1"), ] ensembl_gene_id symbol logFC t P.Value p.adj
7624 ENSG00000149294 NCAM1 12.19755 6.992219 3.81e-08 2.845553e-06Search for 2 genes in the data.frame, CPS1 and GZMB, and verify the effect of adjustment on their p-values
genes <- c("CPS1", "GZMB")
NK_vs_Th |>
filter(symbol %in% genes) |>
select(symbol, P.Value, p.adj) symbol P.Value p.adj
1 CPS1 0.044963086 1.565113e-01
2 GZMB 0.000000629 2.402609e-05CPS1 is not significant, while GZMB is significant.
# Import the adaptive immune response gene set (gmt file)
adaptive <- clusterProfiler::read.gmt("data/GOBP_ADAPTIVE_IMMUNE_RESPONSE.v7.5.1.gmt")
nrow(adaptive) # 719[1] 719length(which(NK_vs_Th$symbol %in% adaptive$gene)) # 513[1] 513upregulated_th <- subset(
NK_vs_Th,
NK_vs_Th$p.adj <= 0.05 & NK_vs_Th$logFC < 0
)
not_significant_genes <- subset(
NK_vs_Th,
NK_vs_Th$p.adj > 0.05
)
summary_upregulated <- summary(upregulated_th$symbol %in% adaptive$gene)
summary_not_significant <- summary(not_significant_genes$symbol %in% adaptive$gene)contingency_table <- matrix(, nrow = 2, ncol = 2)
contingency_table[[1]] <- summary_upregulated[[3]] # up, in gene set
contingency_table[[2]] <- summary_upregulated[[2]] # up, not in gene set
contingency_table[[3]] <- summary_not_significant[[3]] # down, in gene set
contingency_table[[4]] <- summary_not_significant[[2]] # down, not in gene set
# Convert to numeric
contingency_table <- apply(contingency_table, 2, as.numeric)
# Add rows and columns
colnames(contingency_table) <- c("up", "down")
rownames(contingency_table) <- c("in_set", "not_in_set")fisher.test(contingency_table)
Fisher's Exact Test for Count Data
data: contingency_table
p-value < 2.2e-16
alternative hypothesis: true odds ratio is not equal to 1
95 percent confidence interval:
3.697701 5.654348
sample estimates:
odds ratio
4.580549 The odds ratio tells us how different the two proportions are.
If the confidence interval does not include 1, then p-value is small. We can reject null hypothesis thatthe odds ratio is equal to 1.
There are more genes that are upregulated in the gene set than the genes that are not upregulated in the gene set.
# Test 3 gene sets among the genes up-regulated in NK cells,
# with enricher()
# First, obtain the genes up-regulated in NK:
nk_up_genes <- subset(NK_vs_Th, NK_vs_Th$logFC > 0 & NK_vs_Th$p.adj <= 0.05)$symbol
# Import 2 other gene sets, 1 un-related to immune cells:
hair <- read.gmt("data/GOBP_HAIR_CELL_DIFFERENTIATION.v7.5.1.gmt")
dim(hair)[1] 47 2cell_active <- read.gmt("data/GOBP_CELL_ACTIVATION.v7.5.1.gmt")
dim(cell_active)[1] 1095 2# Combine the 3 gene sets into a single data.frame for the TERM2GENE argument:
genesets3 <- rbind(adaptive, hair, cell_active)
hyper_3genesets <- enricher(
gene = nk_up_genes,
universe = NK_vs_Th$symbol,
TERM2GENE = genesets3,
maxGSSize = 1000
)
hyper_3genesets@result ID
GOBP_CELL_ACTIVATION GOBP_CELL_ACTIVATION
GOBP_HAIR_CELL_DIFFERENTIATION GOBP_HAIR_CELL_DIFFERENTIATION
GOBP_ADAPTIVE_IMMUNE_RESPONSE GOBP_ADAPTIVE_IMMUNE_RESPONSE
Description GeneRatio
GOBP_CELL_ACTIVATION GOBP_CELL_ACTIVATION 173/200
GOBP_HAIR_CELL_DIFFERENTIATION GOBP_HAIR_CELL_DIFFERENTIATION 5/200
GOBP_ADAPTIVE_IMMUNE_RESPONSE GOBP_ADAPTIVE_IMMUNE_RESPONSE 82/200
BgRatio pvalue p.adjust qvalue
GOBP_CELL_ACTIVATION 896/1138 0.001505054 0.004515163 0.003168535
GOBP_HAIR_CELL_DIFFERENTIATION 34/1138 0.741306145 0.912609682 0.640427847
GOBP_ADAPTIVE_IMMUNE_RESPONSE 513/1138 0.912609682 0.912609682 0.640427847
geneID
GOBP_CELL_ACTIVATION FGR/CD38/SKAP2/ITGAL/TYROBP/RUNX3/NR1H3/SLAMF7/IFNGR1/STAP1/HDAC9/TSPAN32/RHOA/RAB27A/FCGR2B/TBX21/CST7/HOXA9/ITCH/MEF2C/PTPRC/LAT2/ICAM1/SNAP23/ABL1/HMOX1/IL2RB/SOS2/SLA2/NFATC2/MYL9/NBN/LILRB1/NKG7/CD33/PIK3CG/TGFBR1/MAP3K8/P2RX1/PRKAR1A/CTSC/ZBTB16/CRTAM/TCIRG1/LTBR/MAPK14/PDGFRB/BCL6/CD86/CBLB/LOXL3/IGFBP2/IL18R1/ID2/PLEK/PLA2G4A/RAB29/CD160/RPS6KA1/FOXO3/TNFSF11/PTK2B/SASH3/CD244/BTN2A2/SOX4/IRF1/TNFSF14/RAC2/CRACR2A/BST2/THEMIS2/AP3B1/PRAM1/SWAP70/VAV3/PTPN22/KLRC1/HAVCR2/TEC/DYSF/SERPINE2/LCP1/LRRC8A/TLN1/THBS1/ADAM10/MMRN1/PPP3CA/SLC15A4/IRF8/ERBB2/AKT1/XCL1/ILDR2/SOX13/DGKQ/MSN/GSN/CD226/IL18/DLG5/ADAM8/HHEX/CD8A/SH3RF1/KIT/SLAMF8/F11R/FCER1G/ITGB2/FCRL3/TNFSF13/VCAM1/EOMES/PRKCD/CSF2/SYK/NFIL3/ILK/NFKBID/TMIGD2/CD300A/NLRC3/CCDC88B/PTGDR/CX3CR1/INPP5D/PRELID1/P2RY1/ITGAM/LGALS9B/LGALS9C/AZU1/CD7/PTPN2/GAPT/YES1/ZBTB7A/PRF1/CXCR2/F2R/FES/PLCB1/ADGRG3/MRGPRX2/TBK1/JAG2/CD300LF/FANCA/PLSCR1/CNR2/NCR1/HSH2D/SPN/PLCG2/CARD11/SH2D1B/FCGR3A/NCR3/KLRC2/TRDC/IGHA2/IGHA1/CLIC1/KLRK1/LILRA2/HLA-DMB/DDOST/LYN/KLRC4-KLRK1/DGKK/CCL3
GOBP_HAIR_CELL_DIFFERENTIATION TRIOBP/JAG1/SLC9A3R1/NOTCH1/JAG2
GOBP_ADAPTIVE_IMMUNE_RESPONSE SLAMF7/RAB27A/FCGR2B/PVR/TBX21/STX7/MEF2C/PTPRC/LAT2/ICAM1/SLA2/JAG1/NBN/LILRB1/LILRA1/PIK3CG/CTSC/CRTAM/TCIRG1/TFEB/BCL6/CD86/LOXL3/IL18R1/PLA2G4A/CD160/PTK2B/ADCY7/SASH3/CD244/IRF1/C3/CRACR2A/RFTN1/SWAP70/KLRD1/KLRC1/HAVCR2/TEC/DBNL/SLC15A4/SIGLEC10/XCL1/NOTCH1/CD226/IL18/CD8A/FCER1G/TNFSF13/EOMES/PRKCD/ERAP1/SYK/NFKBID/LAIR1/CX3CR1/INPP5D/CD7/GAPT/C8G/PRF1/SPN/CLEC4C/SH2D1B/FCGR3A/KLRC2/IGLV2-18/IGLV2-11/TRGV9/TRBJ2-1/TRBJ2-2/TRBJ2-3/TRBJ2-4/TRBJ2-7/TRDC/IGHA2/IGHA1/SIPA1/KLRK1/HLA-DMB/LYN/KLRC4-KLRK1
Count
GOBP_CELL_ACTIVATION 173
GOBP_HAIR_CELL_DIFFERENTIATION 5
GOBP_ADAPTIVE_IMMUNE_RESPONSE 82sig_genes <- subset(NK_vs_Th, NK_vs_Th$symbol %in% adaptive$gene &
NK_vs_Th$p.adj <= 0.05)
sig_genes_label <- subset(sig_genes, sig_genes$p.adj <= 0.00001)
ggplot(NK_vs_Th, aes(
x = logFC,
y = -log10(p.adj)
)) +
geom_point(color = "grey87") +
ggtitle("Genes belonging to the adaptive immune response gene set") +
theme_bw() +
geom_text_repel(
data = sig_genes_label,
aes(
x = logFC,
y = -log10(p.adj), label = symbol
),
max.overlaps = 20
) +
geom_point(data = sig_genes, col = "dodgerblue2") +
theme(legend.position = "none") +
scale_x_continuous(name = expression("log"[2] * "(fold change), NK vs Th cells")) +
scale_y_continuous(name = expression("-" * "log"[10] * "(adj. p-value)")) +
geom_hline(yintercept = -log10(0.05), linetype = "dashed") +
geom_vline(xintercept = 0, linetype = "dashed")Warning: ggrepel: 46 unlabeled data points (too many overlaps). Consider
increasing max.overlaps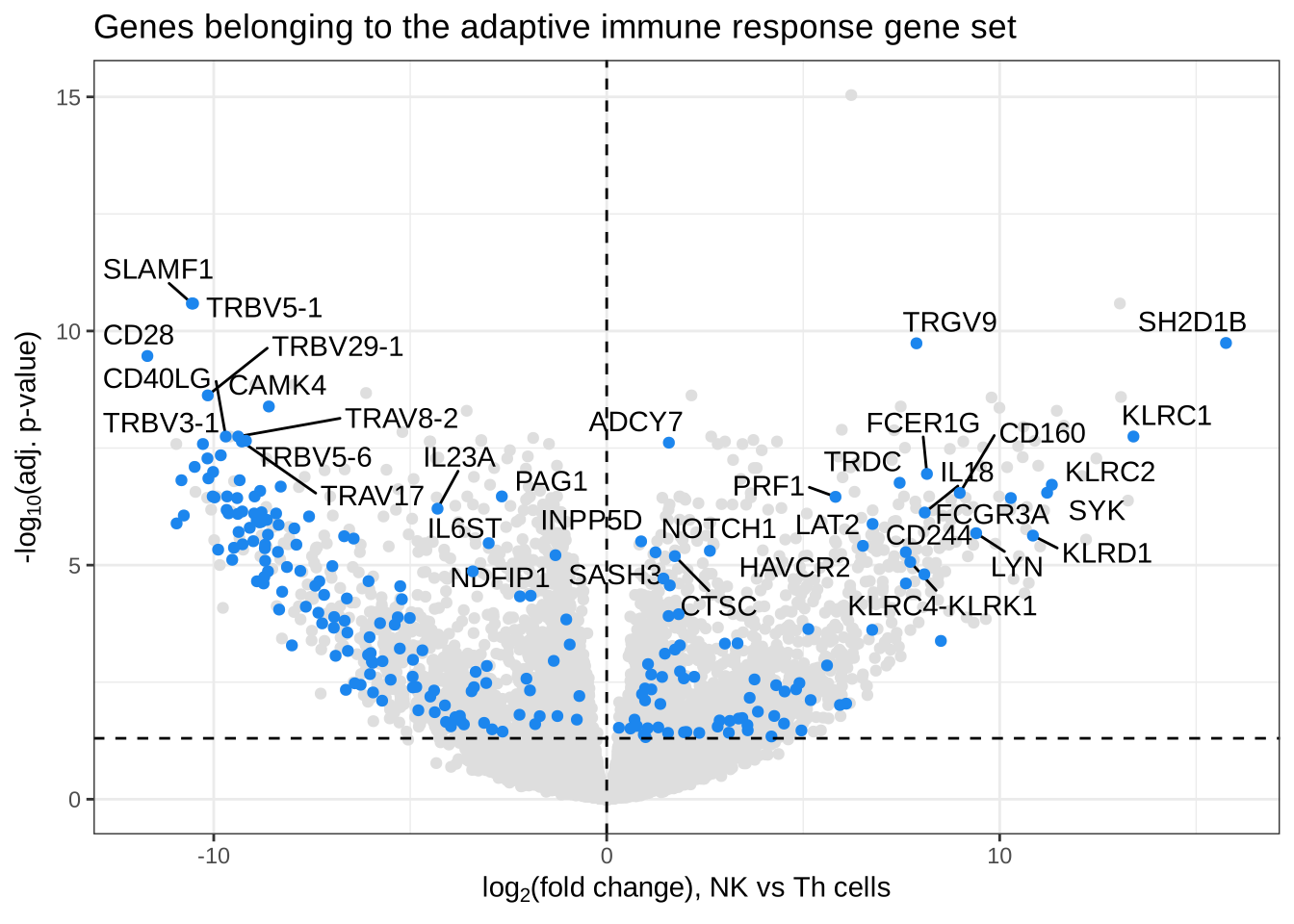
Exercise 2 - Gene set enrichment analysis (GSEA)
gl <- NK_vs_Th$t
names(gl) <- make.names(NK_vs_Th$symbol, unique = T)
gl <- gl[order(gl, decreasing = T)]
GO_NK_Th <- gseGO(gl,
ont = "BP",
OrgDb = org.Hs.eg.db,
keyType = "SYMBOL",
minGSSize = 30,
eps = 0,
seed = T
)preparing geneSet collections...GSEA analysis...Warning in preparePathwaysAndStats(pathways, stats, minSize, maxSize, gseaParam, : There are ties in the preranked stats (0.07% of the list).
The order of those tied genes will be arbitrary, which may produce unexpected results.leading edge analysis...done...GO_NK_Th#
# Gene Set Enrichment Analysis
#
#...@organism Homo sapiens
#...@setType BP
#...@keytype SYMBOL
#...@geneList Named num [1:20485] 19 13.1 12.1 12 10.7 ...
- attr(*, "names")= chr [1:20485] "GHSR" "MLC1" "SH2D1B" "TRGV9" ...
#...nPerm
#...pvalues adjusted by 'BH' with cutoff <0.05
#...351 enriched terms found
'data.frame': 351 obs. of 11 variables:
$ ID : chr "GO:0002181" "GO:0042254" "GO:0022613" "GO:0042273" ...
$ Description : chr "cytoplasmic translation" "ribosome biogenesis" "ribonucleoprotein complex biogenesis" "ribosomal large subunit biogenesis" ...
$ setSize : int 145 299 436 69 98 57 210 340 111 242 ...
$ enrichmentScore: num -0.808 -0.551 -0.491 -0.715 -0.648 ...
$ NES : num -3.38 -2.52 -2.36 -2.67 -2.58 ...
$ pvalue : num 1.68e-48 6.30e-24 2.34e-22 1.48e-14 3.84e-14 ...
$ p.adjust : num 4.85e-45 9.08e-21 2.25e-19 1.07e-11 2.21e-11 ...
$ qvalue : num 3.77e-45 7.05e-21 1.75e-19 8.27e-12 1.72e-11 ...
$ rank : num 1385 3723 3746 2698 2371 ...
$ leading_edge : chr "tags=58%, list=7%, signal=54%" "tags=38%, list=18%, signal=32%" "tags=34%, list=18%, signal=29%" "tags=52%, list=13%, signal=45%" ...
$ core_enrichment: chr "EIF2S2/EIF3M/RPL21/RPS28/EIF4A2/YBX1/FAU/PKM/RPS9/CNBP/RPL37/RPL37A/UBA52/RPS15/RPL22/RPL27/RPS7/RPS11/RPL35/RP"| __truncated__ "UTP23/TMA16/ZNF658/NOB1/WDR43/DHX37/MRPL20/RPS24/EIF2A/ABCE1/RBFA/USP16/METTL16/NOL11/EXOSC8/EXOSC6/TRAF7/LYAR/"| __truncated__ "SRSF6/UTP23/TMA16/ZNF658/SRPK1/NOB1/WDR43/DHX37/MRPL20/RPS24/EIF2A/SF3B5/ABCE1/PTBP2/RBFA/USP16/METTL16/NOL11/S"| __truncated__ "ZNHIT3/SDAD1/MRTO4/RPL26L1/RPLP0P6/FASTKD2/NHP2/NOP16/RSL24D1/PES1/DHX30/WDR74/MALSU1/RSL1D1/NOL9/DDX18/NSA2/GT"| __truncated__ ...
#...Citation
T Wu, E Hu, S Xu, M Chen, P Guo, Z Dai, T Feng, L Zhou, W Tang, L Zhan, X Fu, S Liu, X Bo, and G Yu.
clusterProfiler 4.0: A universal enrichment tool for interpreting omics data.
The Innovation. 2021, 2(3):100141 # Class is gseaResult
class(GO_NK_Th)[1] "gseaResult"
attr(,"package")
[1] "DOSE"# Is the adaptive immune response gene set significant?
GO_NK_Th[GO_NK_Th@result$Description == "adaptive immune response", ] # yes ID Description setSize enrichmentScore
GO:0002250 GO:0002250 adaptive immune response 423 -0.3652034
NES pvalue p.adjust qvalue rank
GO:0002250 -1.743904 1.001619e-08 1.804167e-06 1.400949e-06 1623
leading_edge
GO:0002250 tags=23%, list=8%, signal=22%
core_enrichment
GO:0002250 HFE/CD3E/CLU/PDCD1LG2/ADGRE1/JAK3/LEF1/IL18BP/ITK/CD80/ALCAM/TRAV34/AIRE/IGHM/BTLA/CR1/C1QBP/CD3G/CTSL/TRAJ42/TRBV16/TNF/CEACAM1/GPR183/CD27/CCR6/ICOSLG/TRDV1/CCR2/CD6/TRBD1/MCOLN2/TRAV14DV4/IL2/CR2/TRAV22/CD70/PDCD1/MALT1/EBAG9/TRAV30/CTLA4/TRAV23DV6/TNFRSF13C/KDM5D/TRAV40/TRAV18/IL6R/CD3D/TRAJ3/TRAV39/TRBC2/SAMSN1/IL7R/TRAV19/SUSD4/TRAV20/CD84/TRAV10/TRAV21/TRBV13/TRAV41/TRAV29DV5/NDFIP1/TRAV36DV7/THEMIS/TRBV18/TRAT1/SOCS3/IL6ST/TRBV9/TRAV24/TRAV3/TRAV27/TRAV4/TRAV6/TRAV2/TRAV5/JUNB/TRBV19/TRAV35/TRBV30/FOXP3/TRAV16/IL23A/TRBV2/TRBV14/PAG1/CD4/TRAV25/SIT1/TRAV17/CD40LG/CAMK4/TRAC/CD28/SLAMF1# How many gene sets are down- or up-regulated?
count_gene_sets <- function(gsea, p_value) {
up <- summary(gsea@result$p.adjust < p_value & gsea@result$NES > 0)
down <- summary(gsea@result$p.adjust < p_value & gsea@result$NES < 0)
return(list(upregulated = up, downregulated = down))
}
# 290 upregulated, 61 downregulated
count_gene_sets(GO_NK_Th, 0.05)$upregulated
Mode FALSE TRUE
logical 61 290
$downregulated
Mode FALSE TRUE
logical 290 61 GO_NK_Th_simplify <- clusterProfiler::simplify(GO_NK_Th)
GO_NK_Th_simplify@result[GO_NK_Th_simplify@result$Description == "adaptive immune response", ] ID Description setSize enrichmentScore
GO:0002250 GO:0002250 adaptive immune response 423 -0.3652034
NES pvalue p.adjust qvalue rank
GO:0002250 -1.743904 1.001619e-08 1.804167e-06 1.400949e-06 1623
leading_edge
GO:0002250 tags=23%, list=8%, signal=22%
core_enrichment
GO:0002250 HFE/CD3E/CLU/PDCD1LG2/ADGRE1/JAK3/LEF1/IL18BP/ITK/CD80/ALCAM/TRAV34/AIRE/IGHM/BTLA/CR1/C1QBP/CD3G/CTSL/TRAJ42/TRBV16/TNF/CEACAM1/GPR183/CD27/CCR6/ICOSLG/TRDV1/CCR2/CD6/TRBD1/MCOLN2/TRAV14DV4/IL2/CR2/TRAV22/CD70/PDCD1/MALT1/EBAG9/TRAV30/CTLA4/TRAV23DV6/TNFRSF13C/KDM5D/TRAV40/TRAV18/IL6R/CD3D/TRAJ3/TRAV39/TRBC2/SAMSN1/IL7R/TRAV19/SUSD4/TRAV20/CD84/TRAV10/TRAV21/TRBV13/TRAV41/TRAV29DV5/NDFIP1/TRAV36DV7/THEMIS/TRBV18/TRAT1/SOCS3/IL6ST/TRBV9/TRAV24/TRAV3/TRAV27/TRAV4/TRAV6/TRAV2/TRAV5/JUNB/TRBV19/TRAV35/TRBV30/FOXP3/TRAV16/IL23A/TRBV2/TRBV14/PAG1/CD4/TRAV25/SIT1/TRAV17/CD40LG/CAMK4/TRAC/CD28/SLAMF1unlist(strsplit(
GO_NK_Th@result[GO_NK_Th@result$Description == "adaptive immune response", 11],
"\\/"
)) [1] "HFE" "CD3E" "CLU" "PDCD1LG2" "ADGRE1" "JAK3"
[7] "LEF1" "IL18BP" "ITK" "CD80" "ALCAM" "TRAV34"
[13] "AIRE" "IGHM" "BTLA" "CR1" "C1QBP" "CD3G"
[19] "CTSL" "TRAJ42" "TRBV16" "TNF" "CEACAM1" "GPR183"
[25] "CD27" "CCR6" "ICOSLG" "TRDV1" "CCR2" "CD6"
[31] "TRBD1" "MCOLN2" "TRAV14DV4" "IL2" "CR2" "TRAV22"
[37] "CD70" "PDCD1" "MALT1" "EBAG9" "TRAV30" "CTLA4"
[43] "TRAV23DV6" "TNFRSF13C" "KDM5D" "TRAV40" "TRAV18" "IL6R"
[49] "CD3D" "TRAJ3" "TRAV39" "TRBC2" "SAMSN1" "IL7R"
[55] "TRAV19" "SUSD4" "TRAV20" "CD84" "TRAV10" "TRAV21"
[61] "TRBV13" "TRAV41" "TRAV29DV5" "NDFIP1" "TRAV36DV7" "THEMIS"
[67] "TRBV18" "TRAT1" "SOCS3" "IL6ST" "TRBV9" "TRAV24"
[73] "TRAV3" "TRAV27" "TRAV4" "TRAV6" "TRAV2" "TRAV5"
[79] "JUNB" "TRBV19" "TRAV35" "TRBV30" "FOXP3" "TRAV16"
[85] "IL23A" "TRBV2" "TRBV14" "PAG1" "CD4" "TRAV25"
[91] "SIT1" "TRAV17" "CD40LG" "CAMK4" "TRAC" "CD28"
[97] "SLAMF1" GO_NK_Th@geneSets$`GO:0002250` [1] "ADA" "ADCY7" "AGER" "JAG1" "AHR"
[6] "ALCAM" "ALOX15" "ANXA1" "AIRE" "ARG1"
[11] "ARG2" "ASCL2" "B2M" "BCL3" "BCL6"
[16] "TNFRSF17" "CEACAM1" "PRDM1" "BMX" "BTK"
[21] "C1QBP" "SERPING1" "C1QA" "C1QB" "C1QC"
[26] "C1R" "C1S" "C2" "C3" "C4A"
[31] "C4B" "C4BPA" "C4BPB" "C5" "C6"
[36] "C7" "C8A" "C8B" "C8G" "C9"
[41] "CAMK4" "CD1A" "CD1B" "CD1C" "CD1D"
[46] "CD1E" "CD3D" "CD3E" "CD3G" "CD247"
[51] "CD4" "CD6" "CD7" "CD8A" "CD8B"
[56] "CD8B2" "CD19" "CD27" "CD28" "CD80"
[61] "CD86" "CD40" "CD40LG" "CD70" "CD74"
[66] "CD79A" "CD79B" "CD81" "CTSC" "CLC"
[71] "CLU" "CCR6" "CR1" "CR1L" "CR2"
[76] "CSF2RB" "CSK" "CTLA4" "CTSH" "CTSL"
[81] "CTSS" "CX3CR1" "CD55" "GPR183" "EMP2"
[86] "ADGRE1" "EPHB2" "ERCC1" "PTK2B" "FCER1A"
[91] "FCER1G" "FCER2" "FCGR1A" "FCGR1BP" "FCGR2B"
[96] "FCGR3A" "FGA" "FGB" "FGL1" "FOXJ1"
[101] "MTOR" "FUT7" "FYN" "GATA3" "GNL1"
[106] "MSH6" "GZMM" "NCKAP1L" "HFE" "HLA-A"
[111] "HLA-B" "HLA-C" "HLA-DMA" "HLA-DMB" "HLA-DOA"
[116] "HLA-DOB" "HLA-DPA1" "HLA-DPB1" "HLA-DQA1" "HLA-DQA2"
[121] "HLA-DQB1" "HLA-DQB2" "HLA-DRA" "HLA-DRB1" "HLA-DRB3"
[126] "HLA-DRB4" "HLA-DRB5" "HLA-E" "HLA-F" "HLA-G"
[131] "HLA-H" "MR1" "HLX" "HMGB1" "HPRT1"
[136] "HPX" "HRAS" "HSPD1" "ICAM1" "CFI"
[141] "IFNA1" "IFNA2" "IFNA4" "IFNA5" "IFNA6"
[146] "IFNA7" "IFNA8" "IFNA10" "IFNA13" "IFNA14"
[151] "IFNA16" "IFNA17" "IFNA21" "IFNB1" "IFNG"
[156] "IFNW1" "IGHA1" "IGHA2" "IGHD" "IGHE"
[161] "IGHG1" "IGHG2" "IGHG3" "IGHG4" "IGHM"
[166] "JCHAIN" "IGKC" "IGLC1" "IGLC2" "IGLC3"
[171] "IGLC6" "IGLL1" "IL1B" "IL1R1" "IL2"
[176] "IL2RB" "IL4" "IL4R" "IL6" "IL6R"
[181] "IL6ST" "IL7R" "IL9" "IL9R" "IL10"
[186] "IL12A" "IL12B" "IL12RB1" "IL13RA2" "IL17A"
[191] "IL18" "INPP5D" "IRF1" "IRF4" "IRF7"
[196] "ITK" "JAK1" "JAK2" "JAK3" "JUNB"
[201] "KCNJ8" "KLRC1" "KLRC2" "KLRD1" "LAG3"
[206] "LAIR1" "LIG4" "LTA" "LY9" "LYN"
[211] "SH2D1A" "SMAD7" "MBL2" "CD46" "MEF2C"
[216] "MICB" "MLH1" "MPL" "MSH2" "MYD88"
[221] "NBN" "NFKB2" "NOTCH1" "P2RX7" "PDCD1"
[226] "PHB1" "PIK3CD" "PIK3CG" "PLA2G4A" "PMS2"
[231] "PPP3CB" "PRF1" "PRKCB" "PRKCD" "PKN1"
[236] "PRKCQ" "PRKCZ" "PSG9" "PTPN6" "PTPRC"
[241] "PVR" "NECTIN2" "RAB27A" "RAG1" "RAP1GAP"
[246] "RELB" "TRIM27" "RORA" "RORC" "CCL19"
[251] "XCL1" "SIPA1" "SLAMF1" "SLC11A1" "SPN"
[256] "STAT3" "STAT4" "STAT6" "SUPT6H" "SYK"
[261] "ADAM17" "MAP3K7" "TAP1" "TAP2" "TRA"
[266] "TRAV6" "TRB" "TRGC1" "TRGC2" "TRGV1"
[271] "TRGV2" "TRGV3" "TRGV4" "TRGV5" "TRGV8"
[276] "TRGV9" "TRGV10" "TRGV11" "TEC" "TFE3"
[281] "TFRC" "TGFB1" "TLR4" "TNF" "TNFAIP3"
[286] "TNFRSF1B" "TP53BP1" "TRAF2" "TRAF6" "TSC1"
[291] "TNFSF4" "TXK" "TYK2" "UNG" "WAS"
[296] "LAT2" "NSD2" "ZAP70" "ZP3" "FZD5"
[301] "TFEB" "KDM5D" "EOMES" "STX7" "SKAP1"
[306] "TNFSF13" "TNFRSF14" "RIPK2" "FADD" "TNFRSF11A"
[311] "IL18R1" "CD84" "BCL10" "TNFSF18" "SOCS3"
[316] "RNF8" "EXO1" "EBAG9" "IL1RL1" "SLC22A13"
[321] "IL27RA" "SOCS5" "THOC1" "PARP3" "IL18BP"
[326] "EBI3" "LILRB2" "TCIRG1" "CLEC4M" "BTN3A3"
[331] "RAPGEF3" "MAD2L2" "CLEC10A" "BATF" "CXCL13"
[336] "CD226" "TNFSF13B" "MASP2" "TRAF3IP2" "LILRB1"
[341] "ARID5A" "MALT1" "LILRB5" "LILRB4" "LILRA1"
[346] "LILRB3" "LILRA3" "RIPK3" "RAPGEF4" "BTN3A2"
[351] "BTN3A1" "CD160" "DUSP10" "TREX1" "ZBTB1"
[356] "KLRK1" "PAXIP1" "SWAP70" "RAP1GAP2" "RFTN1"
[361] "ICOSLG" "SIRT1" "TNFRSF13B" "CLCF1" "IL17RA"
[366] "PRKD2" "TMEM98" "LAT" "LAMP3" "SIT1"
[371] "TNFRSF21" "IGKV1-5" "IGHV8-51-1" "IGHV7-81" "IGHV6-1"
[376] "IGHV5-10-1" "IGHV5-51" "IGHV4-38-2" "IGHV4-61" "IGHV4-59"
[381] "IGHV4-39" "IGHV4-34" "IGHV4-31" "IGHV4-30-4" "IGHV4-28"
[386] "IGHV4-4" "IGHV3-38-3" "IGHV3-74" "IGHV3-73" "IGHV3-72"
[391] "IGHV3-66" "IGHV3-64" "IGHV3-53" "IGHV3-49" "IGHV3-48"
[396] "IGHV3-43" "IGHV3-38" "IGHV3-35" "IGHV3-33" "IGHV3-30"
[401] "IGHV3-23" "IGHV3-21" "IGHV3-20" "IGHV3-16" "IGHV3-15"
[406] "IGHV3-13" "IGHV3-11" "IGHV3-9" "IGHV3-7" "IGHV2-70"
[411] "IGHV2-26" "IGHV2-5" "IGHV1-69-2" "IGHV1-38-4" "IGHV1-69"
[416] "IGHV1-58" "IGHV1-45" "IGHV1-24" "IGHV1-18" "IGHV1-8"
[421] "IGHV1-3" "IGHJ1" "IGHD1-1" "TRDV3" "TRDV2"
[426] "TRDV1" "TRDJ1" "TRDD1" "TRDC" "TRBV30"
[431] "TRBV29-1" "TRBV28" "TRBV27" "TRBV25-1" "TRBV24-1"
[436] "TRBV23-1" "TRBV20-1" "TRBV19" "TRBV18" "TRBV17"
[441] "TRBV16" "TRBV14" "TRBV13" "TRBV12-5" "TRBV12-4"
[446] "TRBV12-3" "TRBV11-3" "TRBV11-2" "TRBV11-1" "TRBV10-3"
[451] "TRBV10-2" "TRBV10-1" "TRBV9" "TRBV7-9" "TRBV7-8"
[456] "TRBV7-7" "TRBV7-6" "TRBV7-4" "TRBV7-3" "TRBV7-2"
[461] "TRBV7-1" "TRBV6-9" "TRBV6-8" "TRBV6-7" "TRBV6-6"
[466] "TRBV6-5" "TRBV6-4" "TRBV6-3" "TRBV6-1" "TRBV5-8"
[471] "TRBV5-7" "TRBV5-6" "TRBV5-5" "TRBV5-4" "TRBV5-3"
[476] "TRBV5-1" "TRBV4-3" "TRBV4-2" "TRBV4-1" "TRBV3-1"
[481] "TRBV2" "TRBJ2-7" "TRBJ2-6" "TRBJ2-5" "TRBJ2-4"
[486] "TRBJ2-3" "TRBJ2-2" "TRBJ2-1" "TRBJ1-6" "TRBJ1-5"
[491] "TRBJ1-4" "TRBJ1-3" "TRBJ1-2" "TRBJ1-1" "TRBD1"
[496] "TRBC2" "TRBC1" "TRAV41" "TRAV40" "TRAV39"
[501] "TRAV38-2DV8" "TRAV38-1" "TRAV36DV7" "TRAV35" "TRAV34"
[506] "TRAV30" "TRAV29DV5" "TRAV27" "TRAV26-2" "TRAV26-1"
[511] "TRAV25" "TRAV24" "TRAV23DV6" "TRAV22" "TRAV21"
[516] "TRAV20" "TRAV19" "TRAV18" "TRAV17" "TRAV16"
[521] "TRAV14DV4" "TRAV13-2" "TRAV13-1" "TRAV12-3" "TRAV12-2"
[526] "TRAV12-1" "TRAV10" "TRAV9-2" "TRAV9-1" "TRAV8-6"
[531] "TRAV8-4" "TRAV8-3" "TRAV8-2" "TRAV8-1" "TRAV7"
[536] "TRAV5" "TRAV4" "TRAV3" "TRAV2" "TRAV1-2"
[541] "TRAV1-1" "TRAJ42" "TRAJ31" "TRAJ3" "TRAC"
[546] "IGLV11-55" "IGLV10-54" "IGLV9-49" "IGLV8-61" "IGLV7-46"
[551] "IGLV7-43" "IGLV6-57" "IGLV5-52" "IGLV5-48" "IGLV5-45"
[556] "IGLV5-39" "IGLV5-37" "IGLV4-69" "IGLV4-60" "IGLV4-3"
[561] "IGLV3-32" "IGLV3-27" "IGLV3-25" "IGLV3-22" "IGLV3-21"
[566] "IGLV3-19" "IGLV3-16" "IGLV3-12" "IGLV3-10" "IGLV3-9"
[571] "IGLV3-1" "IGLV2-33" "IGLV2-23" "IGLV2-18" "IGLV2-14"
[576] "IGLV2-11" "IGLV2-8" "IGLV1-51" "IGLV1-50" "IGLV1-47"
[581] "IGLV1-44" "IGLV1-40" "IGLV1-36" "IGLJ1" "IGLC7"
[586] "IGKV6D-41" "IGKV6D-21" "IGKV3D-20" "IGKV3D-15" "IGKV3D-11"
[591] "IGKV3D-7" "IGKV2D-30" "IGKV2D-29" "IGKV2D-28" "IGKV2D-26"
[596] "IGKV2D-24" "IGKV1D-43" "IGKV1D-42" "IGKV1D-39" "IGKV1D-37"
[601] "IGKV1D-33" "IGKV1D-17" "IGKV1D-13" "IGKV1D-12" "IGKV1D-8"
[606] "IGKV6-21" "IGKV5-2" "IGKV4-1" "IGKV3-20" "IGKV3-15"
[611] "IGKV3-7" "IGKV2-40" "IGKV2-30" "IGKV2-29" "IGKV2-28"
[616] "IGKV2-24" "IGKV1-39" "IGKV1-37" "IGKV1-27" "IGKV1-17"
[621] "IGKV1-16" "IGKV1-13" "IGKV1-12" "IGKV1-9" "IGKV1-8"
[626] "IGKV1-6" "IGKJ1" "DBNL" "PYCARD" "CD274"
[631] "TBX21" "CD209" "IL21R" "TRAT1" "CLEC4A"
[636] "FOXP3" "EXOSC3" "ZBTB7B" "KMT5B" "LEF1"
[641] "C1RL" "TLR8" "IL23A" "CYRIB" "CD244"
[646] "ERAP1" "IL20RB" "TREM2" "TREM1" "SASH3"
[651] "SHLD2" "RC3H2" "TRPM4" "LAX1" "LIME1"
[656] "RNF125" "SUSD4" "AKIRIN2" "RIF1" "OTUB1"
[661] "PAG1" "CTNNBL1" "IFNK" "DUSP22" "HMCES"
[666] "OTUD7B" "ENTPD7" "MCOLN1" "IGHV7-4-1" "IGHV3-30-3"
[671] "AICDA" "SLAMF7" "HMHB1" "BACH2" "MYO1G"
[676] "SAMSN1" "NOD2" "ERAP2" "CARD9" "SEMA4A"
[681] "NFKBIZ" "DCLRE1C" "CLEC7A" "LILRA6" "ULBP3"
[686] "VTCN1" "BTNL8" "ATAD5" "SVEP1" "ZC3H12A"
[691] "ULBP2" "ULBP1" "PDCD1LG2" "PRR7" "NDFIP1"
[696] "FBXO38" "UNC93B1" "FCRL4" "JAM3" "FCAMR"
[701] "SLA2" "SANBR" "LOXL3" "CRACR2A" "KMT5C"
[706] "NFKBID" "HAVCR2" "ORAI1" "IGHV3-30-5" "SIGLEC10"
[711] "KLHL6" "IL33" "RSAD2" "CLEC6A" "IL17F"
[716] "NLRP3" "SLAMF6" "TNFRSF13C" "SH2D1B" "EXOSC6"
[721] "SLC15A4" "RNF19B" "RAET1E" "RC3H1" "IL23R"
[726] "SHLD1" "BTLA" "RAET1L" "DENND1B" "RNF168"
[731] "CLEC4C" "APLF" "UNC13D" "GAPT" "MARCHF8"
[736] "IL27" "MCOLN2" "ZNF683" "IL4I1" "NLRP10"
[741] "CLEC4D" "IFNE" "CLEC4G" "RAET1G" "NCR3LG1"
[746] "THEMIS" "MIR21" "EIF2AK4" "TARM1" "SCART1"
[751] "CCR2" "C17orf99" "IGLL5" "MICA" "KLRC4-KLRK1"
[756] "IGHV2-70D" "IGHV1-69D" "IGHV3-64D" "IGHV3-43D" "SHLD3" GO_enrich <- enrichGO(
gene = nk_up_genes,
OrgDb = org.Hs.eg.db,
keyType = "SYMBOL",
ont = "MF", # ont="MF" is the default
minGSSize = 30, universe = NK_vs_Th$symbol
)Exercise 3 - Visualization of enrichment results
par(mar = c(5, 20, 3, 3))
barplot(rev(-log10(GO_NK_Th@result$p.adjust[1:10])),
horiz = T, names = rev(GO_NK_Th@result$Description[1:10]),
las = 2, xlab = "-log10(adj.p-value)",
cex.names = 0.7,
col = "lightgreen"
)
abline(v = -log10(0.05))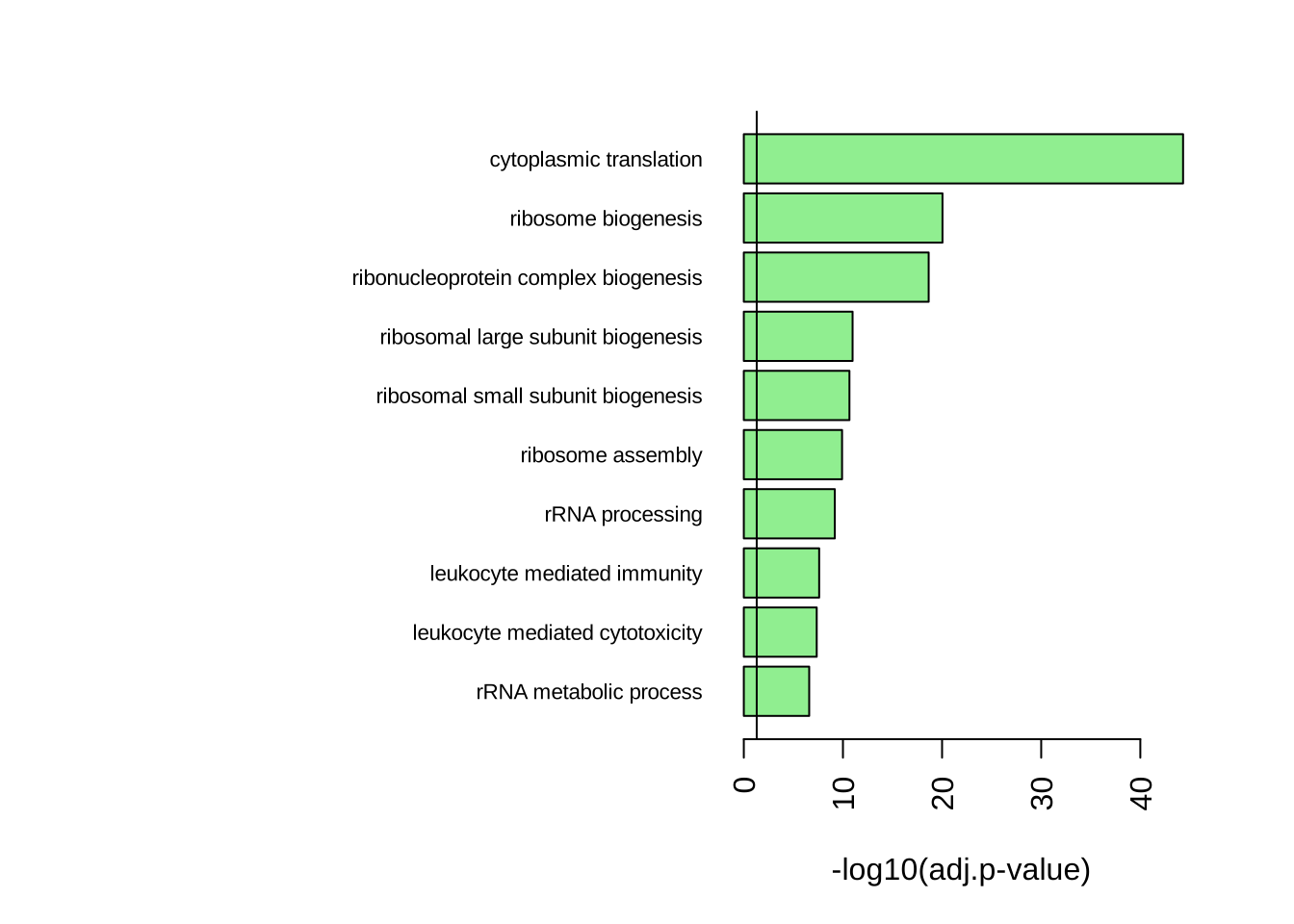
sorted_GO_NK_Th <- GO_NK_Th@result[order(GO_NK_Th@result$NES, decreasing = F), ]
sorted_GO_NK_Th$colors <- ifelse(sorted_GO_NK_Th$NES > 0, "red", "blue")
# Get the indices of the vector
bottom_values <- tail(seq_along(sorted_GO_NK_Th$NES), 10)
par(mar = c(5, 15, 3, 3)) # Make the figure canvas larger
barplot(sorted_GO_NK_Th$NES[c(1:10, bottom_values:nrow(sorted_GO_NK_Th))],
horiz = T, names = sorted_GO_NK_Th$Description[c(1:10, bottom_values:nrow(sorted_GO_NK_Th))],
las = 2, xlab = "NES",
cex.names = 0.7,
col = sorted_GO_NK_Th$color[c(1:10, (nrow(sorted_GO_NK_Th) - 9):nrow(sorted_GO_NK_Th))]
)Warning in bottom_values:nrow(sorted_GO_NK_Th): numerical expression has 10
elements: only the first used
Warning in bottom_values:nrow(sorted_GO_NK_Th): numerical expression has 10
elements: only the first usedabline(v = 0)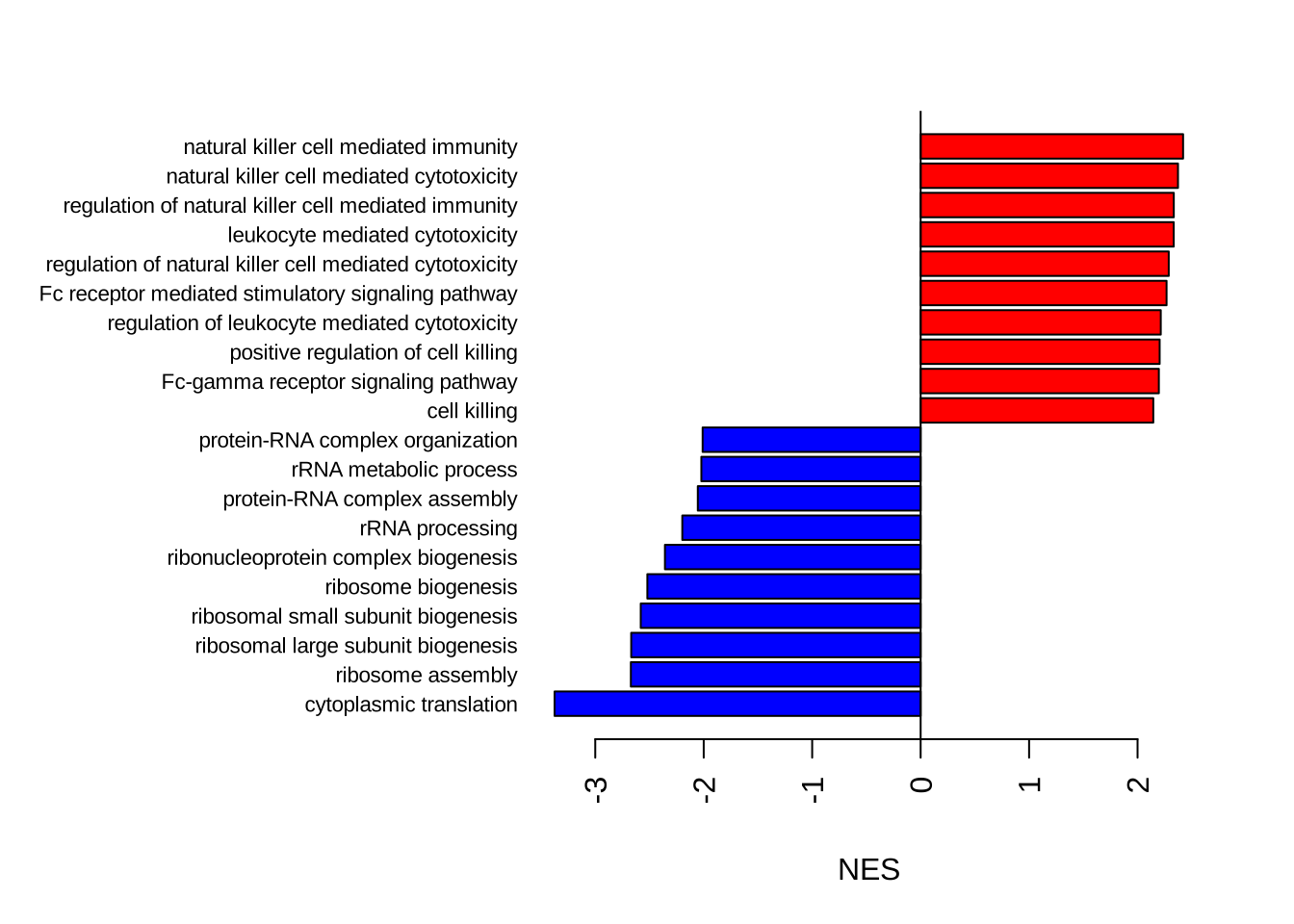
# Use the GO_enrich analysis performed above, of the over-representation analysis
# of genes up-regulated in NK cells:
# barplot() can be directly used on enrichResult objects: but not on gseaResult objects
graphics::barplot(GO_enrich)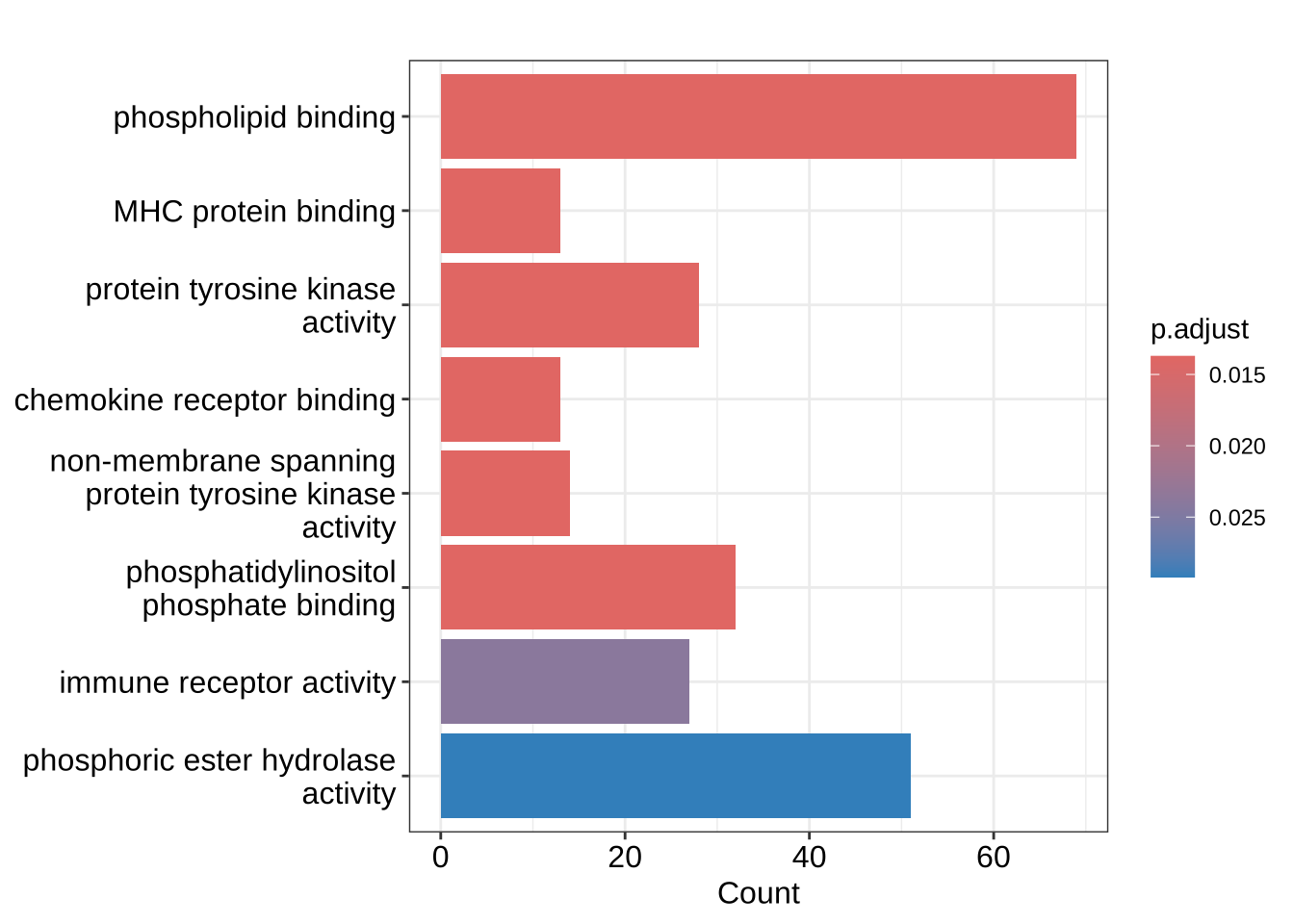
graphics::barplot(GO_enrich, color = "qvalue", x = "GeneRatio")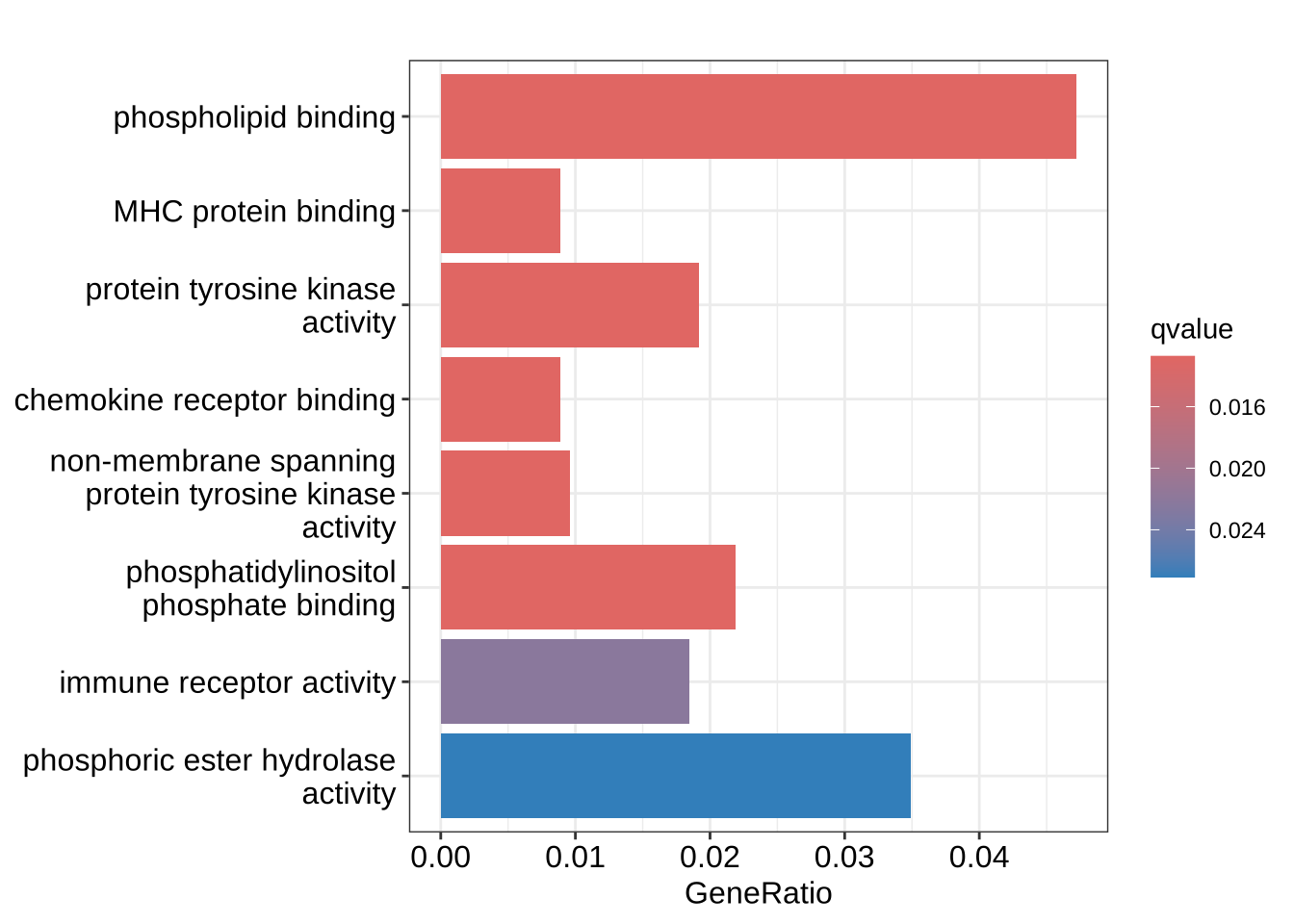
# Select only 2 out of the significant gene sets:
ego_selection <- GO_enrich[GO_enrich@result$ID == "GO:0042287" | GO_enrich@result$ID == "GO:0004713", asis = T]
barplot(ego_selection)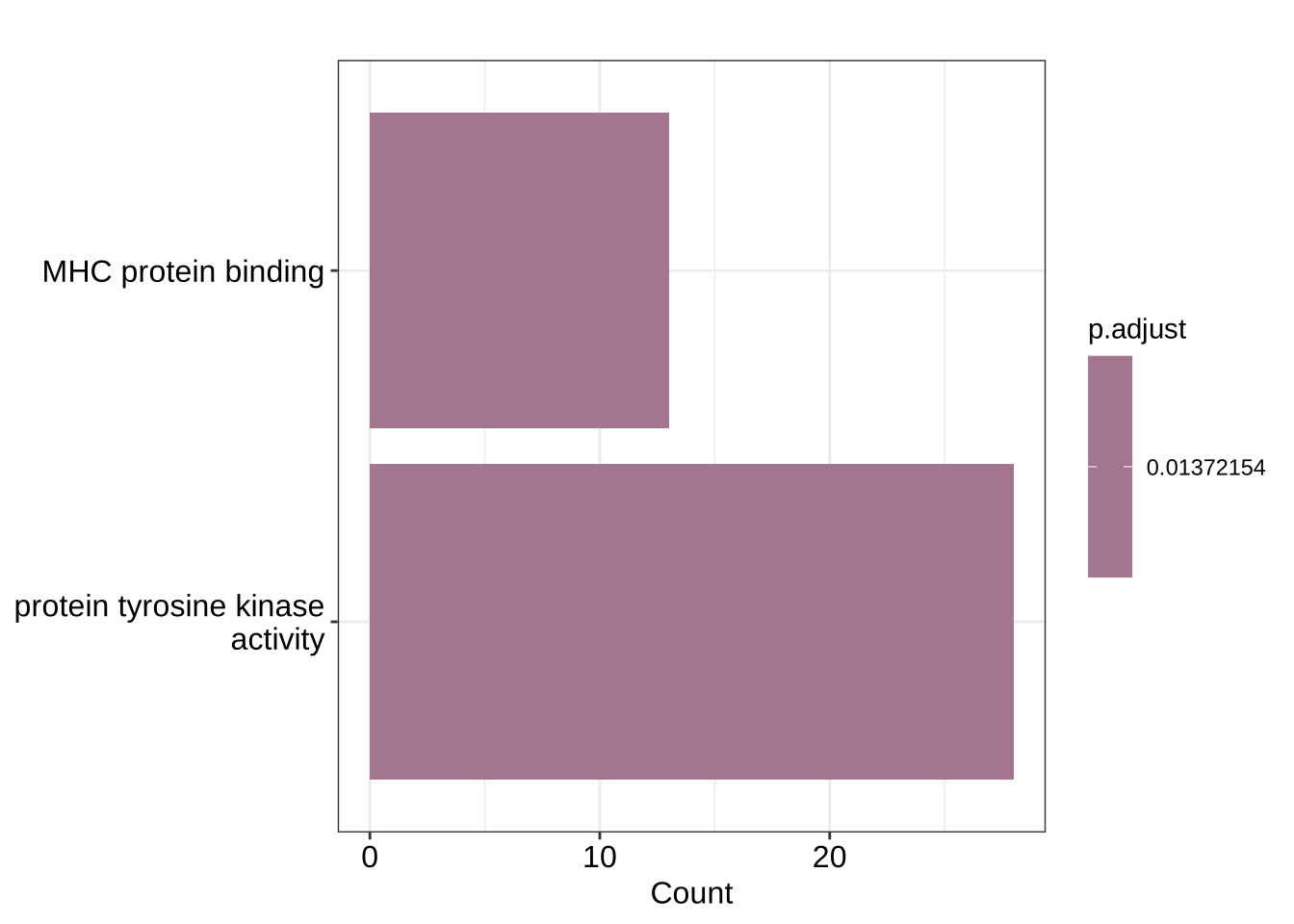
# Barcode plot
# You need the ID of the GO gene set to plot:
GO_NK_Th@result[1:10, 1:6] ID Description setSize
GO:0002181 GO:0002181 cytoplasmic translation 145
GO:0042254 GO:0042254 ribosome biogenesis 299
GO:0022613 GO:0022613 ribonucleoprotein complex biogenesis 436
GO:0042273 GO:0042273 ribosomal large subunit biogenesis 69
GO:0042274 GO:0042274 ribosomal small subunit biogenesis 98
GO:0042255 GO:0042255 ribosome assembly 57
GO:0006364 GO:0006364 rRNA processing 210
GO:0002443 GO:0002443 leukocyte mediated immunity 340
GO:0001909 GO:0001909 leukocyte mediated cytotoxicity 111
GO:0016072 GO:0016072 rRNA metabolic process 242
enrichmentScore NES pvalue
GO:0002181 -0.8083663 -3.375135 1.684522e-48
GO:0042254 -0.5505406 -2.519660 6.298394e-24
GO:0022613 -0.4906673 -2.357458 2.339798e-22
GO:0042273 -0.7146074 -2.668647 1.478487e-14
GO:0042274 -0.6484741 -2.580517 3.835801e-14
GO:0042255 -0.7361084 -2.671760 2.555521e-13
GO:0006364 -0.5004485 -2.197559 1.614149e-12
GO:0002443 0.4169045 1.955354 6.943557e-11
GO:0001909 0.5720092 2.333365 1.381646e-10
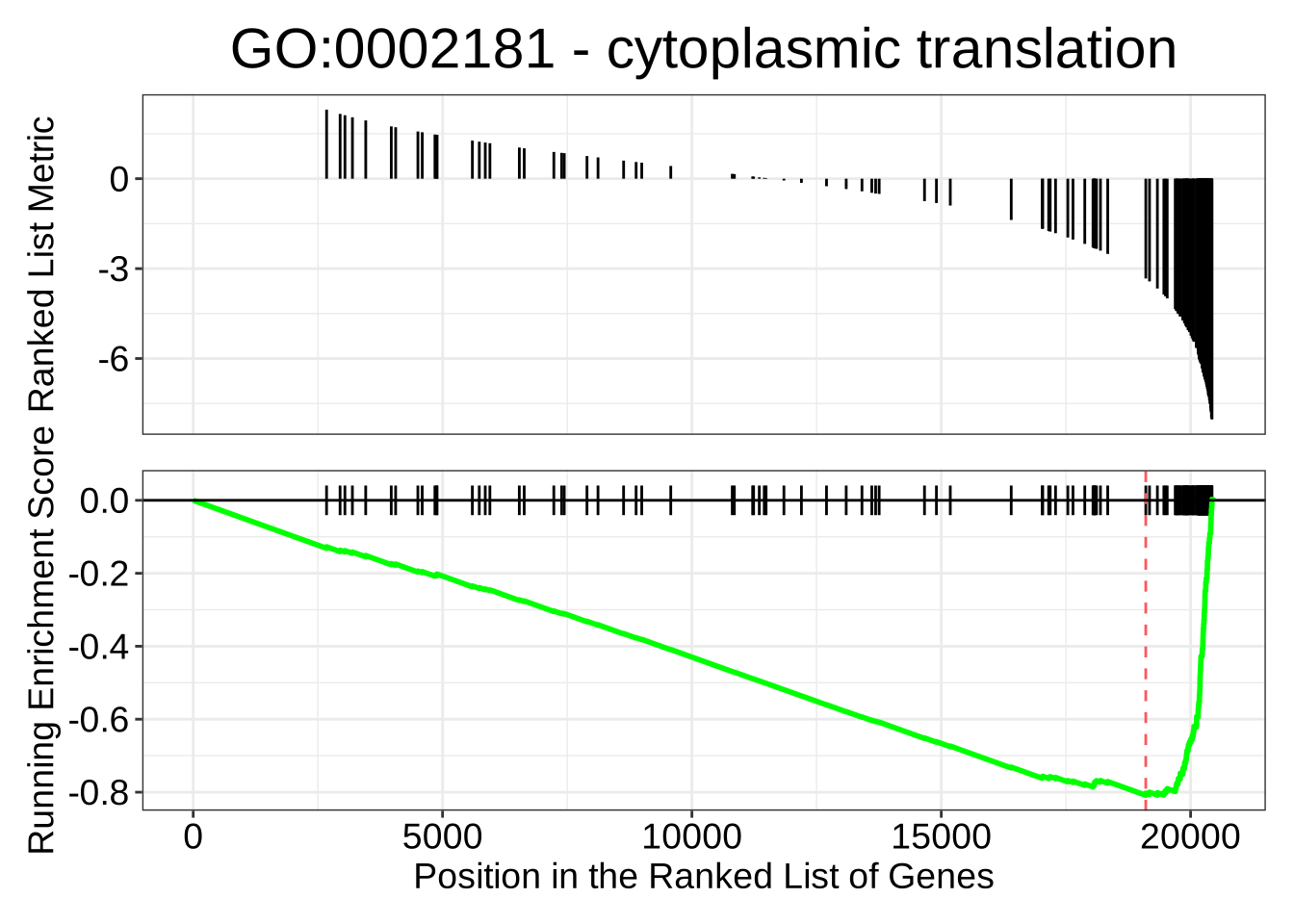
GO:0016072 -0.4491031 -2.021848 8.811745e-10# For a gene set that is down-regulated in NK cells:
gseaplot(GO_NK_Th,
geneSetID = "GO:0002181",
title = "GO:0002181 - cytoplasmic translation"
)
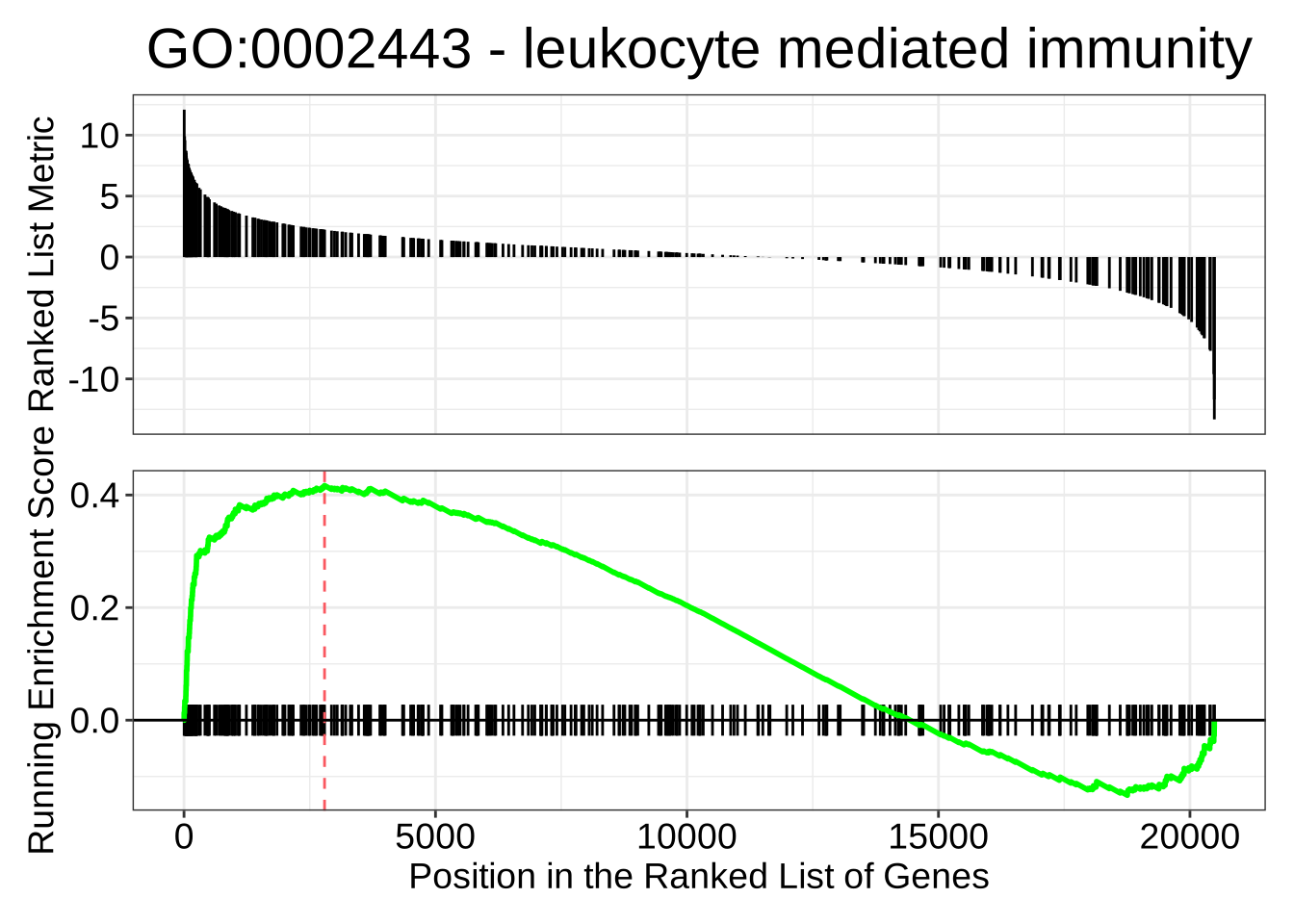
# And one that is up-regulated in NK cells
gseaplot(GO_NK_Th,
geneSetID = "GO:0002443",
title = "GO:0002443 - leukocyte mediated immunity"
)
enrichplot::dotplot(GO_enrich, orderBy = "p.adjust")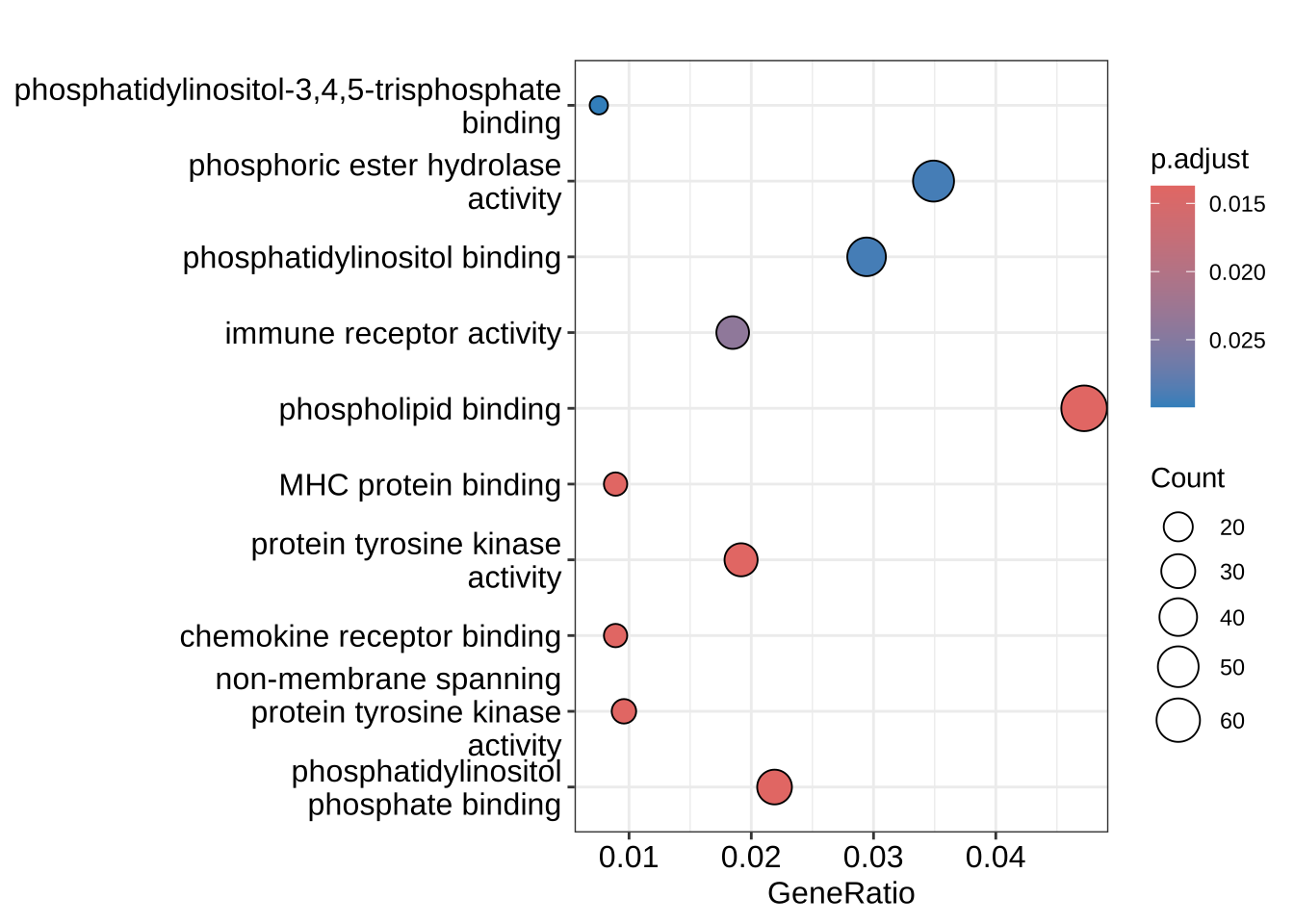
enrichplot::dotplot(GO_NK_Th, orderBy = "p.adjust")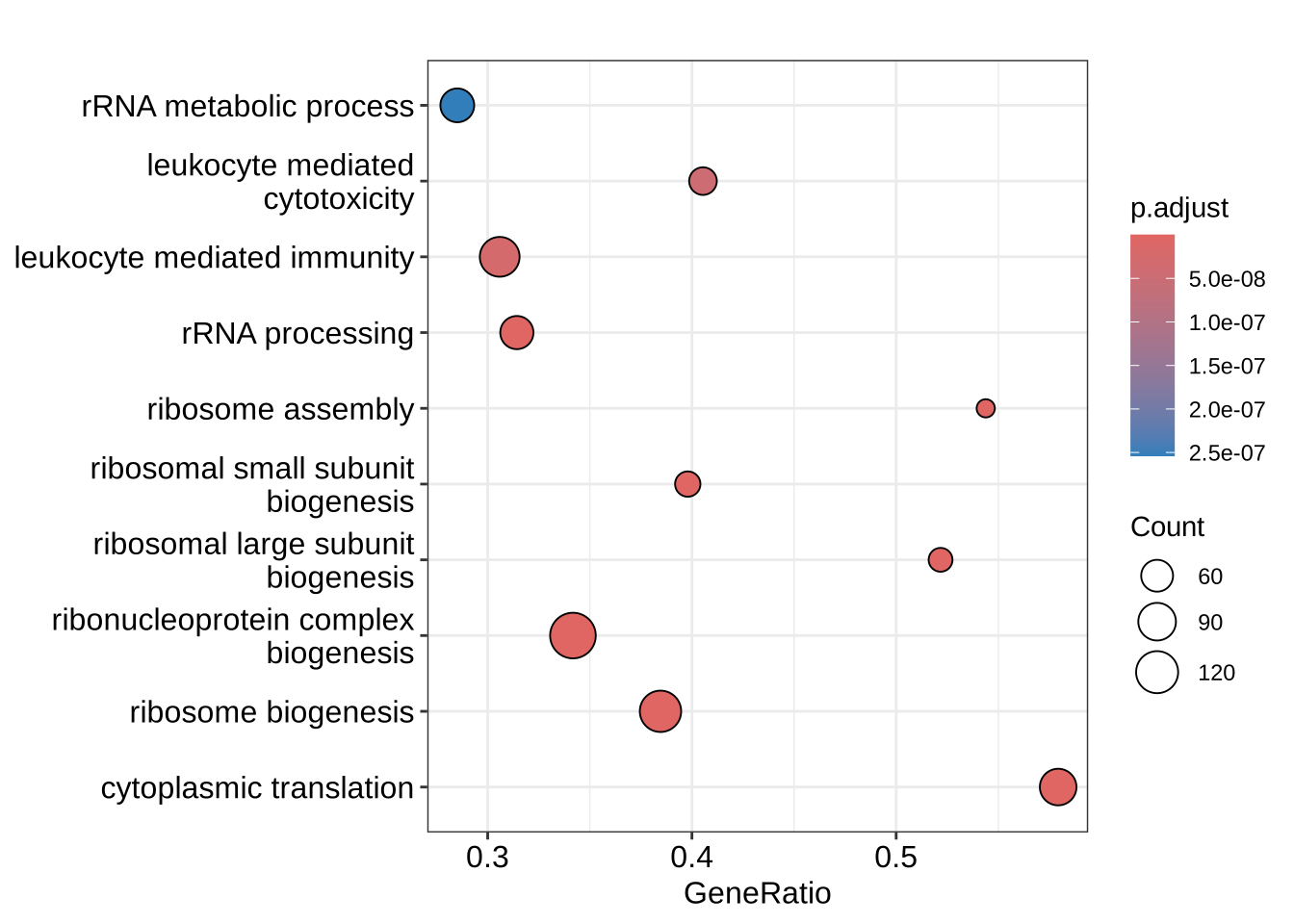
cnetplot(GO_enrich, categorySize = "pvalue")Warning: ggrepel: 68 unlabeled data points (too many overlaps). Consider
increasing max.overlaps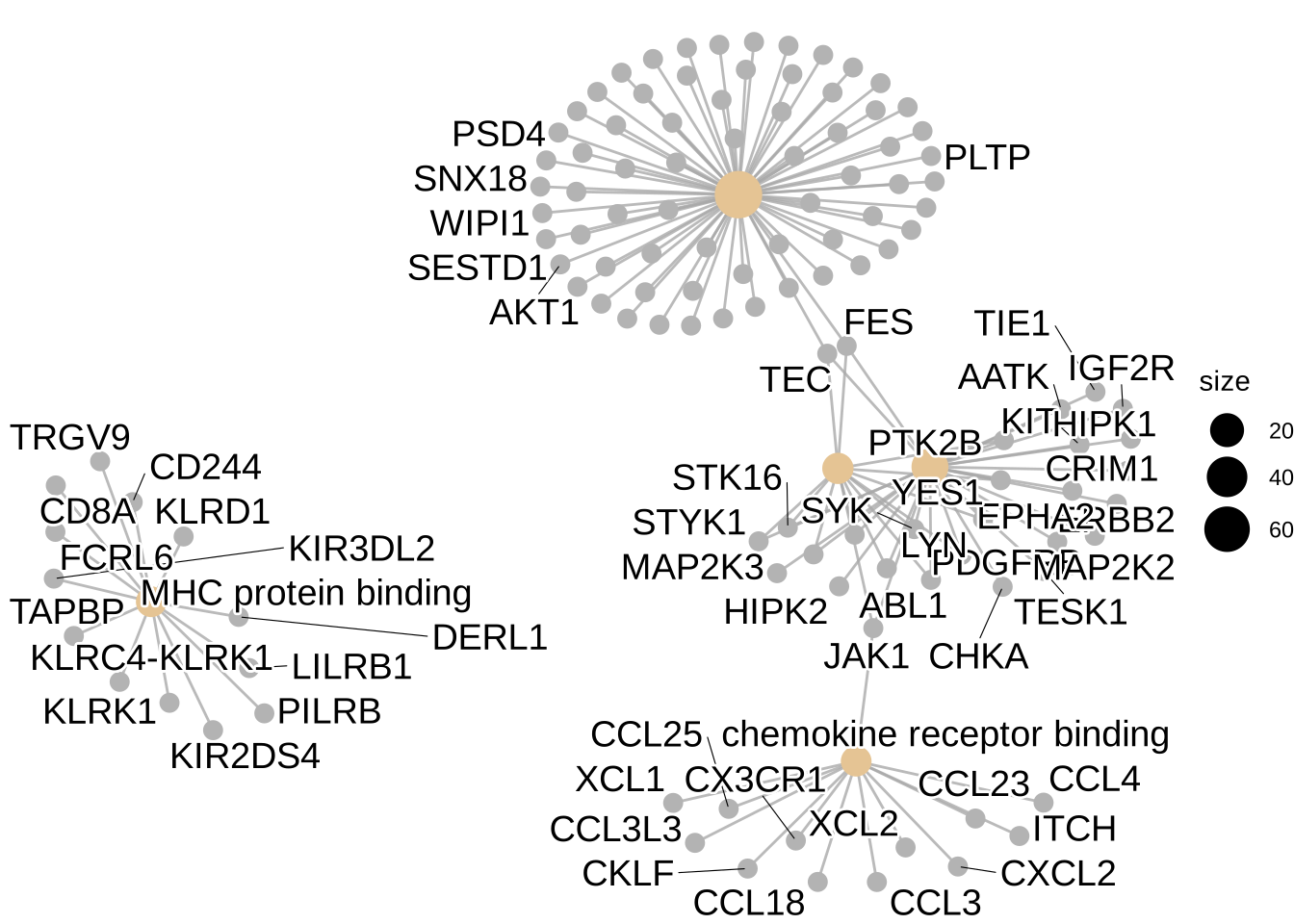
cnetplot(GO_NK_Th, showCategory = 3)Warning: ggrepel: 80 unlabeled data points (too many overlaps). Consider
increasing max.overlaps
ego2 <- pairwise_termsim(GO_NK_Th)
emapplot(ego2, color = "p.adjust")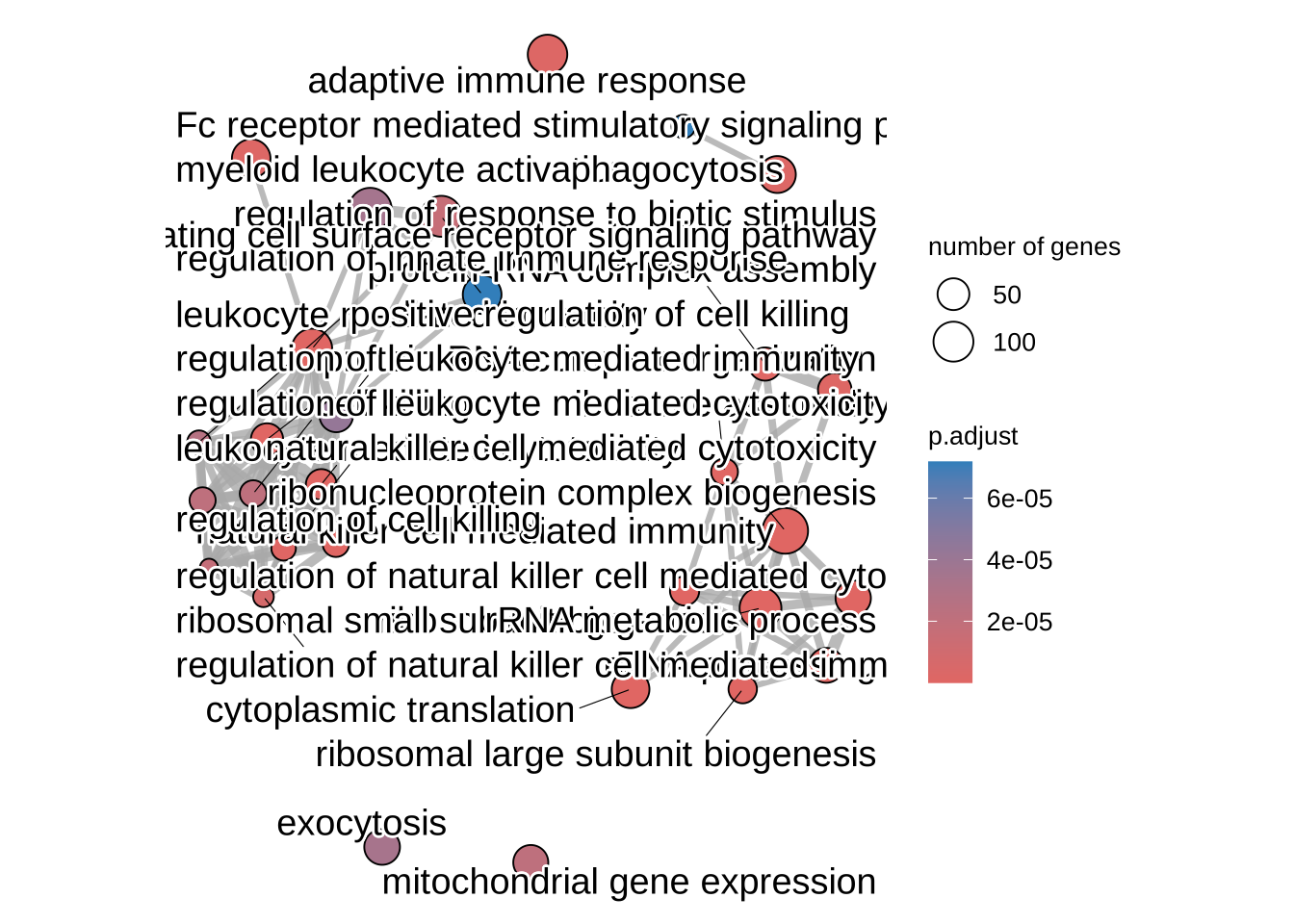
# Wrap lenght of labels
label_format <- 50
# Distribution of t-statistic for genes included in significant gene sets or in selected gene sets:
ridgeplot(GO_NK_Th, label_format = label_format)Picking joint bandwidth of 0.787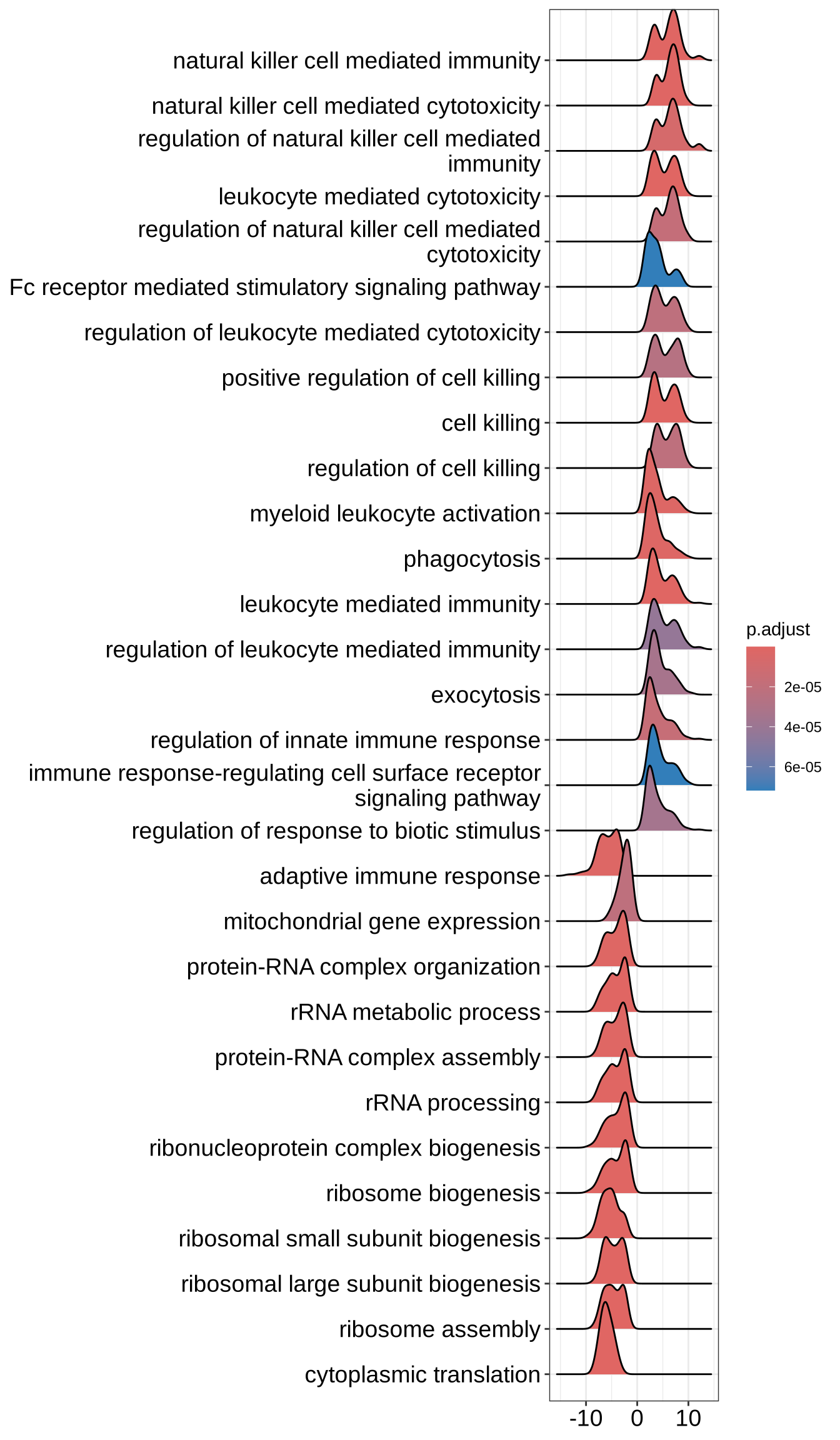
# What is the difference with core_enrichment =F?
ridgeplot(GO_NK_Th, core_enrichment = FALSE, label_format = label_format)Picking joint bandwidth of 0.975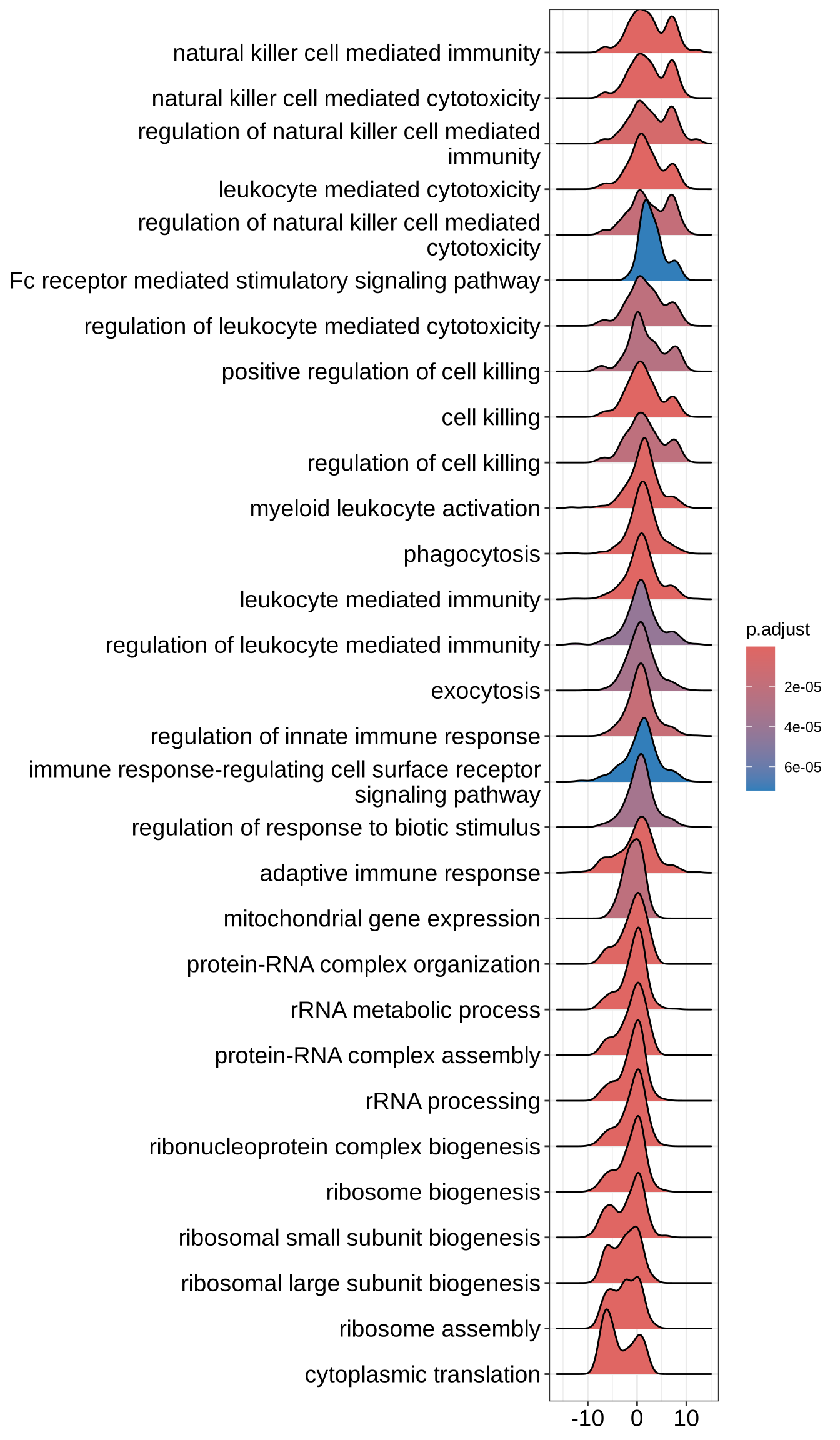
# Select which GO terms to show in the ridge plot:
GO_NK_Th_selection_1 <- GO_NK_Th[GO_NK_Th$ID == "GO:0002181", asis = TRUE]
GO_NK_Th_selection_3 <- GO_NK_Th[
GO_NK_Th$ID %in% c(
"GO:0002181", "GO:0022613",
"GO:0042254"
),
asis = TRUE
]
# Terms that contain the keyword "leukocyte"
GO_NK_Th_selection <- GO_NK_Th[grep("leukocyte", GO_NK_Th@result$Description), asis = TRUE]
ridgeplot(GO_NK_Th_selection, label_format = label_format)Picking joint bandwidth of 0.839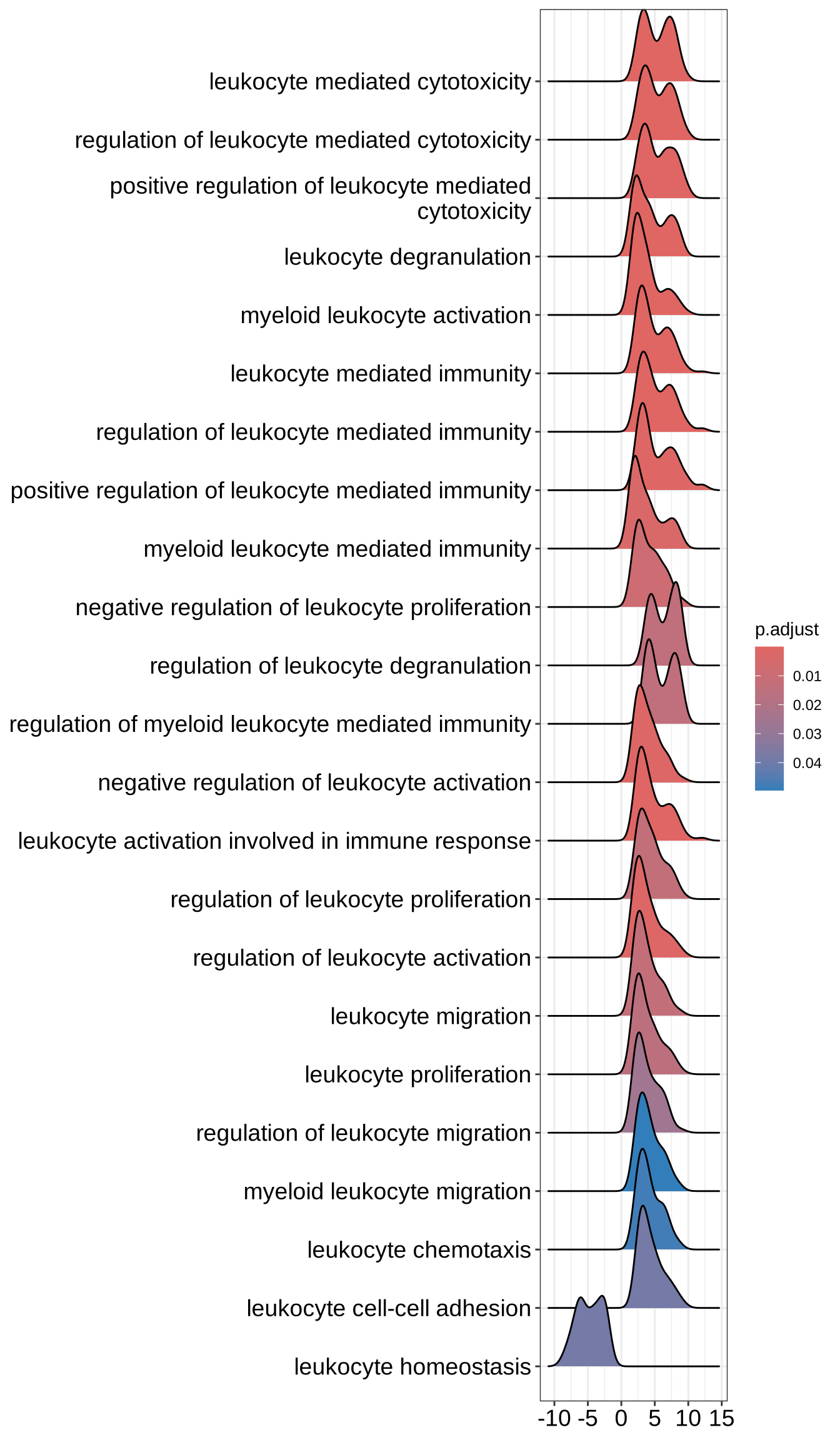
ridgeplot(GO_NK_Th_selection_1)Picking joint bandwidth of 0.423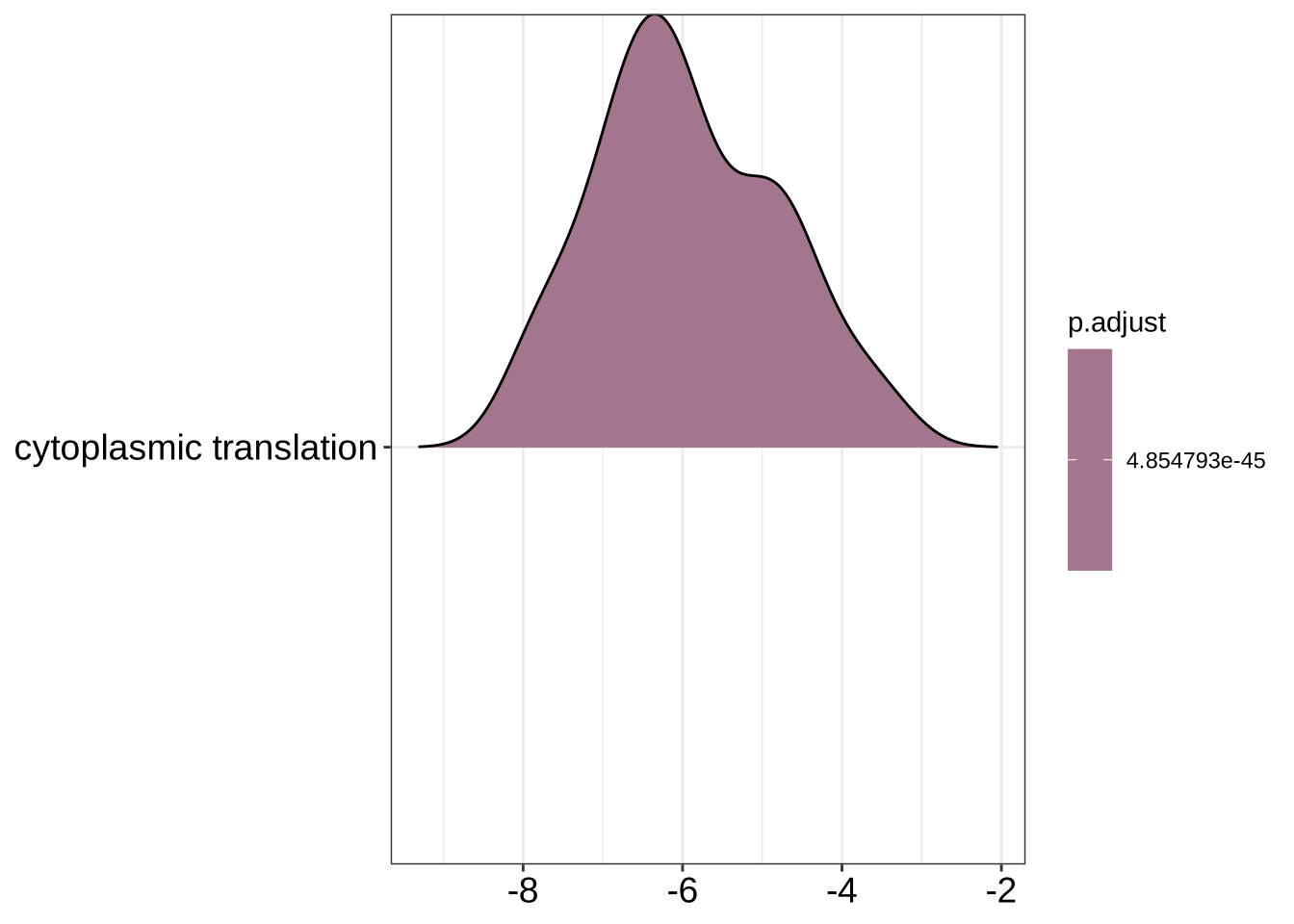
ridgeplot(GO_NK_Th_selection_3)Picking joint bandwidth of 0.589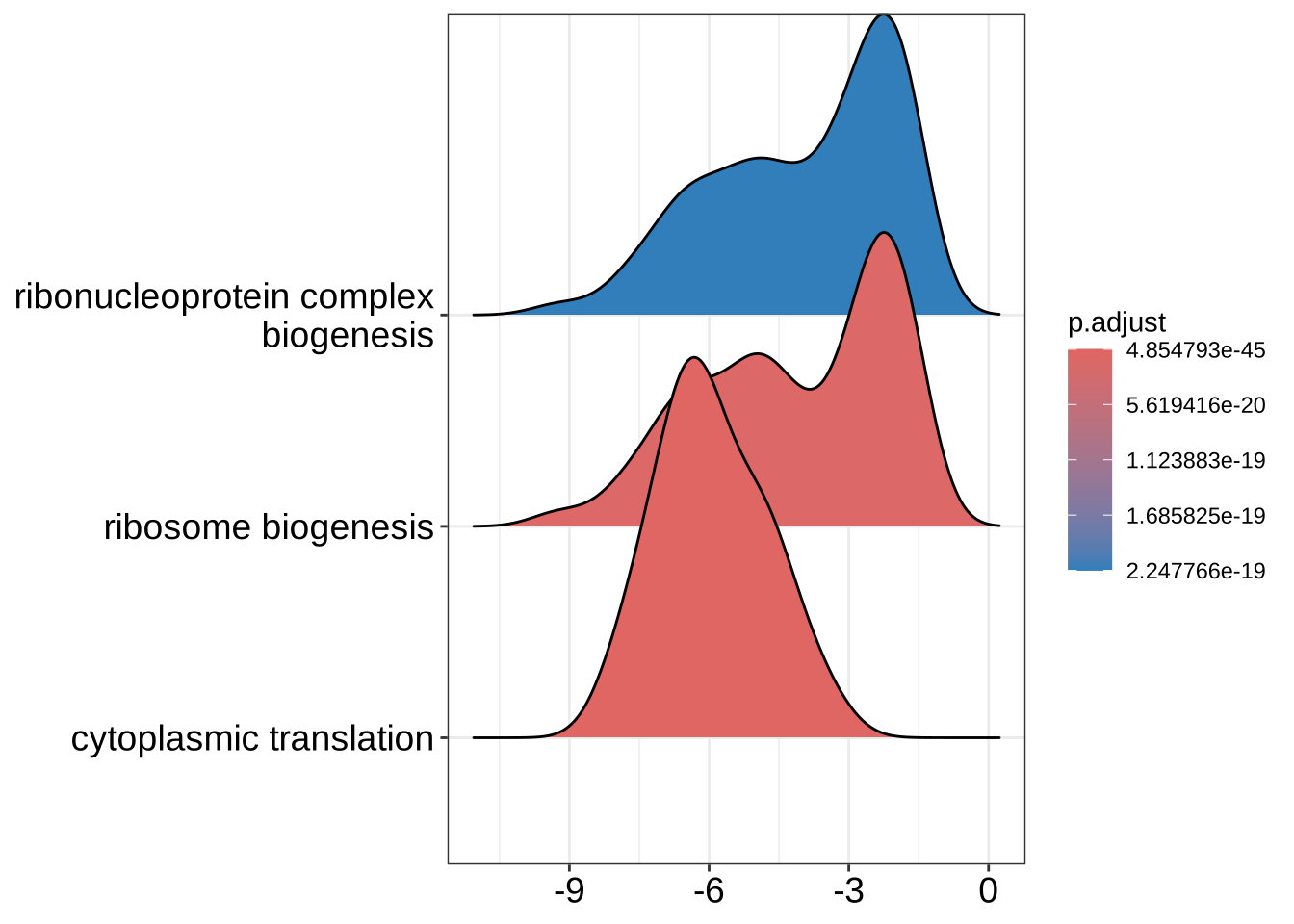
Exercise 4 - Enrichment of other collections of gene sets
keytypes(org.Hs.eg.db) [1] "ACCNUM" "ALIAS" "ENSEMBL" "ENSEMBLPROT" "ENSEMBLTRANS"
[6] "ENTREZID" "ENZYME" "EVIDENCE" "EVIDENCEALL" "GENENAME"
[11] "GENETYPE" "GO" "GOALL" "IPI" "MAP"
[16] "OMIM" "ONTOLOGY" "ONTOLOGYALL" "PATH" "PFAM"
[21] "PMID" "PROSITE" "REFSEQ" "SYMBOL" "UCSCKG"
[26] "UNIPROT" # convert from= "ENSEMBL" to "SYMBOL" and "ENTREZID"
gene_convert <- bitr(as.character(NK_vs_Th$ensembl_gene_id),
fromType = "ENSEMBL",
toType = c("SYMBOL", "ENTREZID"), OrgDb = "org.Hs.eg.db"
)'select()' returned 1:many mapping between keys and columnsWarning in bitr(as.character(NK_vs_Th$ensembl_gene_id), fromType = "ENSEMBL", :
18.73% of input gene IDs are fail to map...# Check the format of the data frame obtained after conversion:
head(gene_convert) ENSEMBL SYMBOL ENTREZID
1 ENSG00000000003 TSPAN6 7105
2 ENSG00000000419 DPM1 8813
3 ENSG00000000457 SCYL3 57147
4 ENSG00000000460 FIRRM 55732
5 ENSG00000000938 FGR 2268
6 ENSG00000000971 CFH 3075dim(gene_convert)[1] 16794 3# Create a vector of genes that are coded with the EntrezID:
# use the sorted gene list gl previously created:
gl_kegg <- cbind(SYMBOL = names(gl), t = gl)
# merge with converted gene symbols to combine both:
# by default the data frames are merged on the columns with names they both have
gl_kegg <- merge(gl_kegg, gene_convert)
head(gl_kegg) SYMBOL t ENSEMBL ENTREZID
1 A1BG 1.129187394 ENSG00000121410 1
2 A2M -0.382294217 ENSG00000175899 2
3 A4GALT 0.808365644 ENSG00000128274 53947
4 AAAS 0.749990903 ENSG00000094914 8086
5 AACS 2.172253591 ENSG00000081760 65985
6 AADAT 3.038354213 ENSG00000109576 51166gl_kegg_list <- as.numeric(as.character(gl_kegg$t))
names(gl_kegg_list) <- as.character(gl_kegg$ENTREZID)
gl_kegg_list <- sort(gl_kegg_list, decreasing = T)
# run GSEA of KEGG (please note that requires internet connection to download the KEGG annotations from http)
KEGG_NK_Th <- gseKEGG(gl_kegg_list,
organism = "hsa", "ncbi-geneid",
minGSSize = 30,
eps = 0,
seed = T
)Reading KEGG annotation online: "https://rest.kegg.jp/link/hsa/pathway"...Reading KEGG annotation online: "https://rest.kegg.jp/list/pathway/hsa"...Reading KEGG annotation online: "https://rest.kegg.jp/conv/ncbi-geneid/hsa"...preparing geneSet collections...GSEA analysis...Warning in preparePathwaysAndStats(pathways, stats, minSize, maxSize, gseaParam, : There are ties in the preranked stats (0.1% of the list).
The order of those tied genes will be arbitrary, which may produce unexpected results.Warning in preparePathwaysAndStats(pathways, stats, minSize, maxSize,
gseaParam, : There are duplicate gene names, fgsea may produce unexpected
results.leading edge analysis...done...# What does it contain?
str(KEGG_NK_Th)Formal class 'gseaResult' [package "DOSE"] with 13 slots
..@ result :'data.frame': 24 obs. of 11 variables:
.. ..$ ID : chr [1:24] "hsa03010" "hsa05171" "hsa04650" "hsa04666" ...
.. ..$ Description : chr [1:24] "Ribosome" "Coronavirus disease - COVID-19" "Natural killer cell mediated cytotoxicity" "Fc gamma R-mediated phagocytosis" ...
.. ..$ setSize : int [1:24] 130 185 98 86 163 181 93 77 118 220 ...
.. ..$ enrichmentScore: num [1:24] -0.813 -0.678 0.62 0.525 0.426 ...
.. ..$ NES : num [1:24] -3.46 -3.02 2.49 2.07 1.87 ...
.. ..$ pvalue : num [1:24] 3.32e-46 5.30e-32 2.03e-12 1.23e-06 2.91e-06 ...
.. ..$ p.adjust : num [1:24] 8.94e-44 7.13e-30 1.82e-10 8.24e-05 1.57e-04 ...
.. ..$ qvalue : num [1:24] 7.31e-44 5.84e-30 1.49e-10 6.74e-05 1.28e-04 ...
.. ..$ rank : num [1:24] 1852 1168 1873 1924 2216 ...
.. ..$ leading_edge : chr [1:24] "tags=72%, list=13%, signal=63%" "tags=48%, list=8%, signal=45%" "tags=52%, list=13%, signal=46%" "tags=42%, list=13%, signal=36%" ...
.. ..$ core_enrichment: chr [1:24] "63875/140032/51121/64983/6139/9553/51021/51116/6133/51065/65008/51187/64960/64981/6166/54460/6144/6234/51081/21"| __truncated__ "5600/6166/3454/3661/5595/6144/6234/7124/5335/7187/2197/9641/6203/6167/6168/7311/3558/6209/6146/6155/6201/6205/1"| __truncated__ "117157/5336/3804/3821/2207/3822/6850/6452/3802/5551/2214/7305/3805/3811/9437/7462/3809/3824/3823/51744/5594/300"| __truncated__ "5336/6850/2214/4067/3635/2934/5594/3984/81873/5880/10451/2212/7408/10092/1785/207/382/2215/2213/1793/8503/5580/"| __truncated__ ...
..@ organism : chr "hsa"
..@ setType : chr "KEGG"
..@ geneSets :List of 366
.. ..$ hsa00010: chr [1:67] "10327" "124" "125" "126" ...
.. ..$ hsa00020: chr [1:30] "1431" "1737" "1738" "1743" ...
.. ..$ hsa00030: chr [1:31] "132158" "2203" "221823" "226" ...
.. ..$ hsa00040: chr [1:36] "10327" "10720" "10941" "231" ...
.. ..$ hsa00051: chr [1:34] "197258" "2203" "226" "229" ...
.. ..$ hsa00052: chr [1:32] "130589" "231" "2538" "2548" ...
.. ..$ hsa00053: chr [1:30] "10327" "10720" "10941" "217" ...
.. ..$ hsa00061: chr [1:18] "109703458" "197322" "2180" "2181" ...
.. ..$ hsa00062: chr [1:28] "10449" "10965" "11332" "117145" ...
.. ..$ hsa00071: chr [1:43] "10449" "10455" "113612" "124" ...
.. ..$ hsa00100: chr [1:20] "1056" "10682" "120227" "1591" ...
.. ..$ hsa00120: chr [1:17] "10005" "10858" "10998" "1109" ...
.. ..$ hsa00130: chr [1:12] "10229" "154807" "1728" "2677" ...
.. ..$ hsa00140: chr [1:63] "100861540" "10720" "10941" "1109" ...
.. ..$ hsa00190: chr [1:138] "100532726" "10063" "101927180" "10312" ...
.. ..$ hsa00220: chr [1:23] "100526760" "1373" "137362" "162417" ...
.. ..$ hsa00230: chr [1:128] "100" "100526794" "10201" "102157402" ...
.. ..$ hsa00232: chr [1:6] "10" "1544" "1548" "1549" ...
.. ..$ hsa00240: chr [1:58] "100526794" "10201" "115024" "124583" ...
.. ..$ hsa00250: chr [1:37] "122622" "1373" "137362" "158" ...
.. ..$ hsa00260: chr [1:41] "102724560" "10993" "113675" "124908081" ...
.. ..$ hsa00270: chr [1:52] "102724560" "1036" "10768" "10993" ...
.. ..$ hsa00280: chr [1:48] "10449" "11112" "1629" "1738" ...
.. ..$ hsa00290: chr [1:4] "10993" "113675" "586" "587"
.. ..$ hsa00310: chr [1:63] "10157" "10919" "11105" "123688" ...
.. ..$ hsa00330: chr [1:50] "112483" "112817" "112849" "113451" ...
.. ..$ hsa00340: chr [1:22] "10841" "131669" "138199" "144193" ...
.. ..$ hsa00350: chr [1:36] "124" "125" "126" "127" ...
.. ..$ hsa00360: chr [1:16] "137362" "1644" "218" "221" ...
.. ..$ hsa00380: chr [1:42] "11185" "121278" "125061" "130013" ...
.. ..$ hsa00400: chr [1:6] "137362" "259307" "2805" "2806" ...
.. ..$ hsa00410: chr [1:31] "18" "1806" "1807" "1892" ...
.. ..$ hsa00430: chr [1:17] "102724197" "1036" "124975" "2326" ...
.. ..$ hsa00440: chr [1:6] "10390" "5130" "56994" "5833" ...
.. ..$ hsa00450: chr [1:17] "10587" "11185" "114112" "118672" ...
.. ..$ hsa00470: chr [1:6] "1610" "27165" "2744" "63826" ...
.. ..$ hsa00480: chr [1:59] "102724197" "10314" "119391" "124975" ...
.. ..$ hsa00500: chr [1:40] "11181" "124905666" "124905668" "128966568" ...
.. ..$ hsa00510: chr [1:55] "10195" "10905" "11253" "11282" ...
.. ..$ hsa00511: chr [1:18] "10825" "129807" "175" "23324" ...
.. ..$ hsa00512: chr [1:36] "100528030" "10331" "10610" "11226" ...
.. ..$ hsa00513: chr [1:43] "10195" "10905" "11253" "11282" ...
.. ..$ hsa00514: chr [1:47] "100528030" "10585" "11226" "11227" ...
.. ..$ hsa00515: chr [1:23] "10329" "10585" "10690" "11041" ...
.. ..$ hsa00520: chr [1:38] "10007" "10020" "1118" "132789" ...
.. ..$ hsa00524: chr [1:5] "2645" "3098" "3099" "3101" ...
.. ..$ hsa00531: chr [1:19] "10855" "138050" "23553" "2588" ...
.. ..$ hsa00532: chr [1:21] "10090" "11285" "113189" "126792" ...
.. ..$ hsa00533: chr [1:14] "10164" "10678" "2530" "2683" ...
.. ..$ hsa00534: chr [1:24] "11285" "126792" "2131" "2132" ...
.. ..$ hsa00541: chr [1:20] "10020" "123956252" "140838" "1727" ...
.. ..$ hsa00561: chr [1:65] "10327" "10554" "10555" "1056" ...
.. ..$ hsa00562: chr [1:73] "10423" "113026" "138429" "200576" ...
.. ..$ hsa00563: chr [1:30] "10026" "128869" "23556" "27315" ...
.. ..$ hsa00564: chr [1:103] "100137049" "10162" "10390" "1040" ...
.. ..$ hsa00565: chr [1:50] "100137049" "10390" "11145" "122618" ...
.. ..$ hsa00590: chr [1:63] "100137049" "102724197" "10728" "11145" ...
.. ..$ hsa00591: chr [1:30] "100137049" "11145" "123745" "151056" ...
.. ..$ hsa00592: chr [1:26] "100137049" "11145" "123745" "151056" ...
.. ..$ hsa00600: chr [1:54] "10558" "10715" "10825" "123099" ...
.. ..$ hsa00601: chr [1:28] "10317" "10331" "10402" "10678" ...
.. ..$ hsa00603: chr [1:16] "10317" "10690" "127550" "2523" ...
.. ..$ hsa00604: chr [1:15] "256435" "2583" "27090" "2720" ...
.. ..$ hsa00620: chr [1:47] "10327" "10873" "124" "125" ...
.. ..$ hsa00630: chr [1:32] "112817" "124908081" "125061" "132158" ...
.. ..$ hsa00640: chr [1:32] "160287" "1629" "1738" "18" ...
.. ..$ hsa00650: chr [1:27] "116285" "123876" "142827" "18" ...
.. ..$ hsa00670: chr [1:39] "100528021" "102724560" "10588" "10768" ...
.. ..$ hsa00730: chr [1:15] "122481" "158067" "203" "204" ...
.. ..$ hsa00740: chr [1:8] "5167" "5169" "52" "53" ...
.. ..$ hsa00750: chr [1:6] "29968" "316" "493911" "55163" ...
.. ..$ hsa00760: chr [1:38] "100526794" "10135" "133686" "22933" ...
.. ..$ hsa00770: chr [1:21] "1806" "1807" "217" "219" ...
.. ..$ hsa00780: chr [1:3] "3141" "54995" "686"
.. ..$ hsa00785: chr [1:20] "11019" "116285" "124908081" "1629" ...
.. ..$ hsa00790: chr [1:28] "10243" "121278" "1719" "200895" ...
.. ..$ hsa00830: chr [1:68] "100861540" "10170" "10720" "10901" ...
.. ..$ hsa00860: chr [1:46] "10720" "10941" "124454" "1352" ...
.. ..$ hsa00900: chr [1:23] "100529261" "10269" "10654" "116150" ...
.. ..$ hsa00910: chr [1:17] "11238" "1373" "23632" "2746" ...
.. ..$ hsa00920: chr [1:10] "10380" "23474" "4357" "54928" ...
.. ..$ hsa00970: chr [1:66] "10056" "10352" "10667" "118672" ...
.. ..$ hsa00980: chr [1:79] "10720" "107987478" "107987479" "10941" ...
.. ..$ hsa00982: chr [1:73] "10720" "107987478" "107987479" "10941" ...
.. ..$ hsa00983: chr [1:81] "10" "10201" "1066" "10720" ...
.. ..$ hsa01040: chr [1:27] "10965" "11332" "122970" "201562" ...
.. ..$ hsa01100: chr [1:1570] "10" "100" "10005" "10007" ...
.. ..$ hsa01200: chr [1:116] "10873" "10993" "113675" "124908081" ...
.. ..$ hsa01210: chr [1:33] "100526760" "137362" "1431" "162417" ...
.. ..$ hsa01212: chr [1:57] "10449" "109703458" "126129" "1374" ...
.. ..$ hsa01230: chr [1:75] "100526760" "102724560" "10993" "113675" ...
.. ..$ hsa01232: chr [1:85] "100" "100526794" "10201" "102157402" ...
.. ..$ hsa01240: chr [1:154] "10201" "102157402" "10229" "10243" ...
.. ..$ hsa01250: chr [1:37] "10020" "140838" "197258" "23483" ...
.. ..$ hsa01320: chr [1:2] "9060" "9061"
.. ..$ hsa01521: chr [1:80] "10000" "10018" "110117499" "1950" ...
.. ..$ hsa01522: chr [1:99] "10000" "1019" "1026" "1027" ...
.. ..$ hsa01523: chr [1:30] "10057" "10257" "113235" "1147" ...
.. ..$ hsa01524: chr [1:75] "10000" "1026" "1029" "110117499" ...
.. .. [list output truncated]
..@ geneList : Named num [1:14284] 19 13.1 12.1 12 10.7 ...
.. ..- attr(*, "names")= chr [1:14284] "2693" "23209" "117157" "6983" ...
..@ keytype : chr "ncbi-geneid"
..@ permScores : num[0 , 0 ]
..@ params :List of 6
.. ..$ pvalueCutoff : num 0.05
.. ..$ eps : num 0
.. ..$ pAdjustMethod: chr "BH"
.. ..$ exponent : num 1
.. ..$ minGSSize : num 30
.. ..$ maxGSSize : num 500
..@ gene2Symbol: chr(0)
..@ readable : logi FALSE
..@ termsim : num[0 , 0 ]
..@ method : chr(0)
..@ dr : list()# How many gene sets are up-regulated?
sum(KEGG_NK_Th@result$NES > 0) # 17[1] 17#|
grep_kegg_description <- function(pattern) {
return(grep(pattern, tolower((KEGG_NK_Th@result$Description))))
}# Is their an immune-related gene set significant?
grep_kegg_description("immune")integer(0)# Is their an NK gene set significant?
grep_kegg_description("natural killer") # 3[1] 3# What is the total number of built-in KEGG gene sets?
length(KEGG_NK_Th@geneSets) # 265[1] 366KEGG_NK_Th[grep_kegg_description("natural killer"), ] |>
select(ID, Description) # hsa04650 ID Description
hsa04650 hsa04650 Natural killer cell mediated cytotoxicityKEGG_NK_Th@geneSets$hsa04650 [1] "100132285" "100507436" "100528032" "102723407" "10451" "10870"
[7] "110117499" "117157" "124905743" "135250" "1437" "154064"
[13] "2185" "2207" "2214" "2215" "22914" "2534"
[19] "25759" "259197" "27040" "2885" "3002" "3105"
[25] "3106" "3107" "3133" "3135" "3265" "3383"
[31] "3384" "3439" "3440" "3441" "3442" "3443"
[37] "3444" "3445" "3446" "3447" "3448" "3449"
[43] "3451" "3452" "3454" "3455" "3456" "3458"
[49] "3459" "3460" "353091" "355" "356" "3683"
[55] "3689" "369" "3802" "3803" "3804" "3805"
[61] "3806" "3808" "3809" "3810" "3811" "3812"
[67] "3821" "3822" "3823" "3824" "3845" "3932"
[73] "3937" "399694" "4068" "4277" "4772" "4773"
[79] "4893" "5058" "51744" "5290" "5291" "5293"
[85] "5295" "5296" "5335" "53358" "5336" "5530"
[91] "5532" "5533" "5534" "5535" "5551" "5578"
[97] "5579" "5582" "5594" "5595" "5604" "5605"
[103] "57292" "5777" "5781" "5879" "5880" "5881"
[109] "5894" "637" "6452" "6464" "6654" "6655"
[115] "673" "6850" "7124" "7305" "7409" "7410"
[121] "7462" "7535" "79465" "80328" "80329" "836"
[127] "8503" "8743" "8795" "8797" "919" "9436"
[133] "9437" "962" # pathview map with non-significant genes in grey:
# set log fold change of non-significant genes to 0:
NK_vs_Th$logFC_0 <- ifelse(NK_vs_Th$p.adj > 0.05, 0, NK_vs_Th$logFC)
# create named vector of fold change values:
genePW <- NK_vs_Th$logFC_0
names(genePW) <- NK_vs_Th$symbol
# Create pathview map for Ribosome = hsa03010
pathview(
gene.data = genePW,
pathway.id = "hsa03010",
species = "hsa",
gene.idtype = "SYMBOL"
)'select()' returned 1:many mapping between keys and columns[1] "Note: 4806 of 20411 unique input IDs unmapped."'select()' returned 1:1 mapping between keys and columnsInfo: Working in directory /var/home/artur/Documents/10-19_PhD/11_Education/11.22-sib-enrichment-analysisInfo: Writing image file hsa03010.pathview.png# Create pathview map of Natural killer cell mediated cytotoxicity = hsa04650
pathview(
gene.data = genePW,
pathway.id = "hsa04650",
species = "hsa",
gene.idtype = "SYMBOL"
)'select()' returned 1:many mapping between keys and columns[1] "Note: 4806 of 20411 unique input IDs unmapped."'select()' returned 1:1 mapping between keys and columnsInfo: Working in directory /var/home/artur/Documents/10-19_PhD/11_Education/11.22-sib-enrichment-analysisInfo: Writing image file hsa04650.pathview.png# Import hallmark, convert to term2gene and run GSEA:
term2gene_h <- msigdbr(species = "Homo sapiens", category = "H")
# Or alternatively:
# term2gene_h<-read.gmt("h.all.v2023.2.Hs.symbols.gmt")
head(term2gene_h)# A tibble: 6 × 15
gs_cat gs_subcat gs_name gene_symbol entrez_gene ensembl_gene
<chr> <chr> <chr> <chr> <int> <chr>
1 H "" HALLMARK_ADIPOGENESIS ABCA1 19 ENSG00000165029
2 H "" HALLMARK_ADIPOGENESIS ABCB8 11194 ENSG00000197150
3 H "" HALLMARK_ADIPOGENESIS ACAA2 10449 ENSG00000167315
4 H "" HALLMARK_ADIPOGENESIS ACADL 33 ENSG00000115361
5 H "" HALLMARK_ADIPOGENESIS ACADM 34 ENSG00000117054
6 H "" HALLMARK_ADIPOGENESIS ACADS 35 ENSG00000122971
# ℹ 9 more variables: human_gene_symbol <chr>, human_entrez_gene <int>,
# human_ensembl_gene <chr>, gs_id <chr>, gs_pmid <chr>, gs_geoid <chr>,
# gs_exact_source <chr>, gs_url <chr>, gs_description <chr>length(unique(term2gene_h$gs_name)) # 50[1] 50# Run GSEA with the function that allows to use custom gene sets,
# provide the named vector of t statistics
h_NK_vs_Th <- GSEA(gl,
TERM2GENE = term2gene_h[, c("gs_name", "gene_symbol")],
eps = 0,
seed = T
)preparing geneSet collections...GSEA analysis...Warning in preparePathwaysAndStats(pathways, stats, minSize, maxSize, gseaParam, : There are ties in the preranked stats (0.07% of the list).
The order of those tied genes will be arbitrary, which may produce unexpected results.leading edge analysis...done...# Number of significant gene sets:
length(which(h_NK_vs_Th@result$p.adjust <= 0.05))[1] 4# A dotplot with geneRatio or NES on the x-axis:
dotplot(h_NK_vs_Th)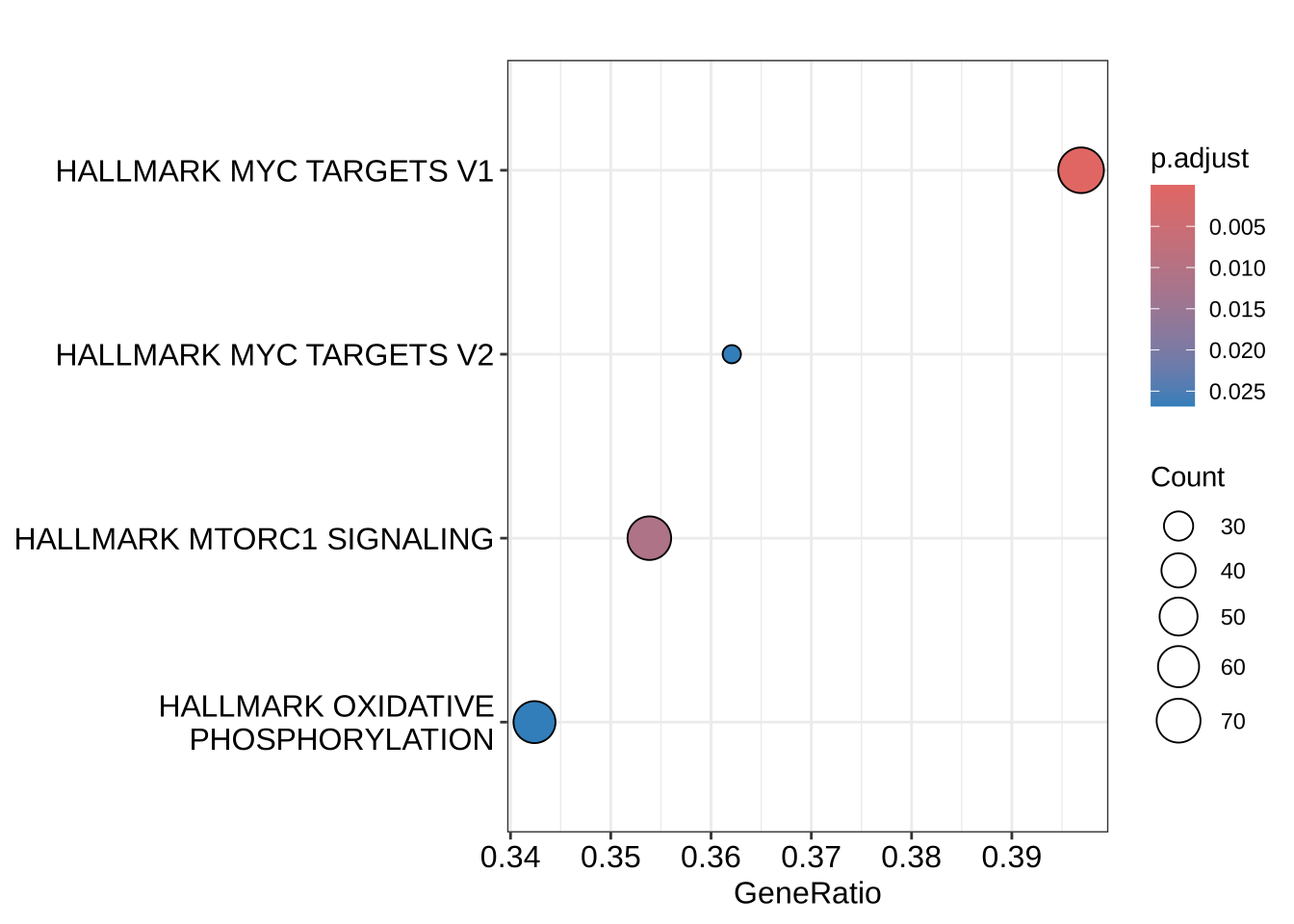
dotplot(h_NK_vs_Th, x = "NES", orderBy = "p.adjust")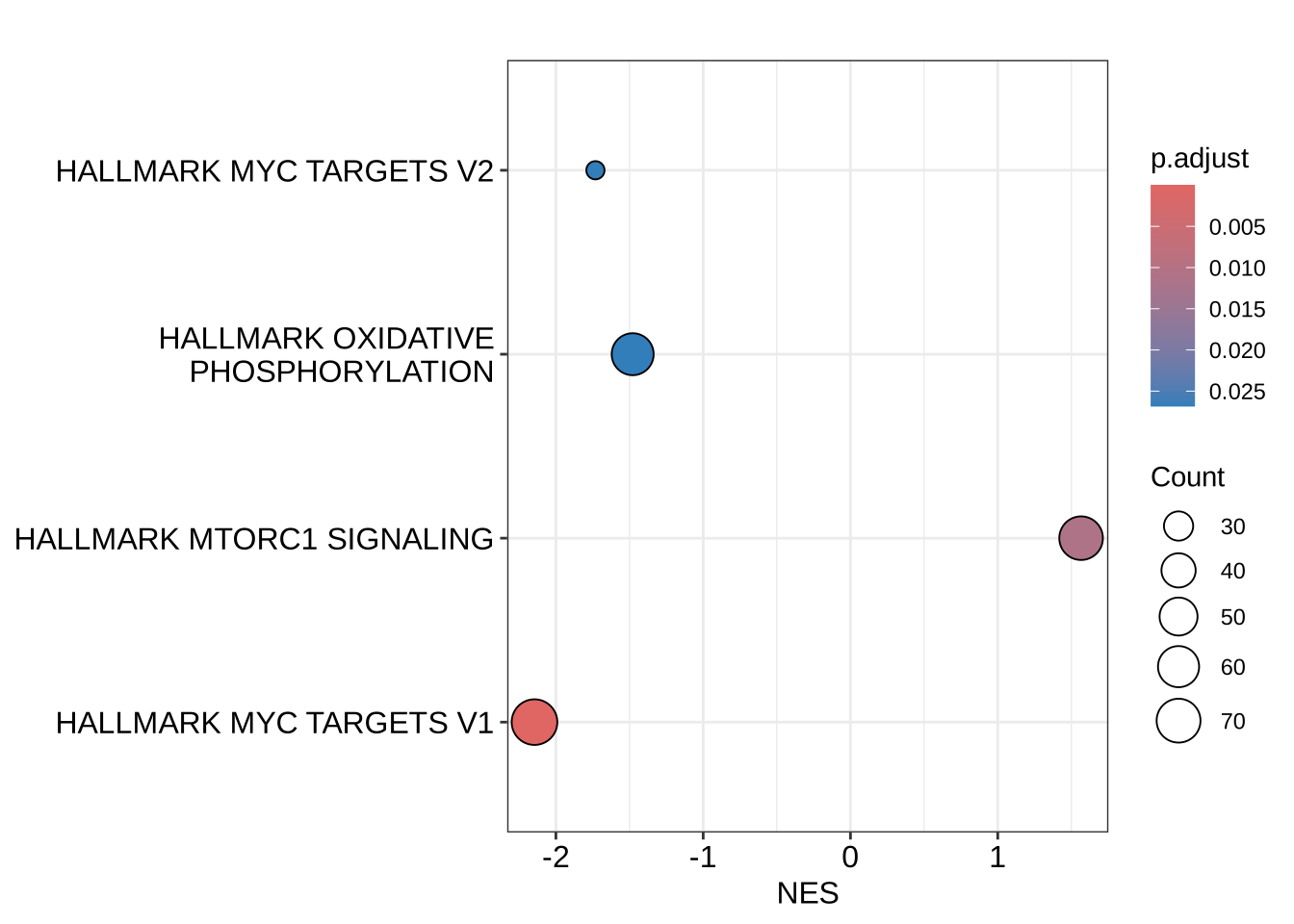
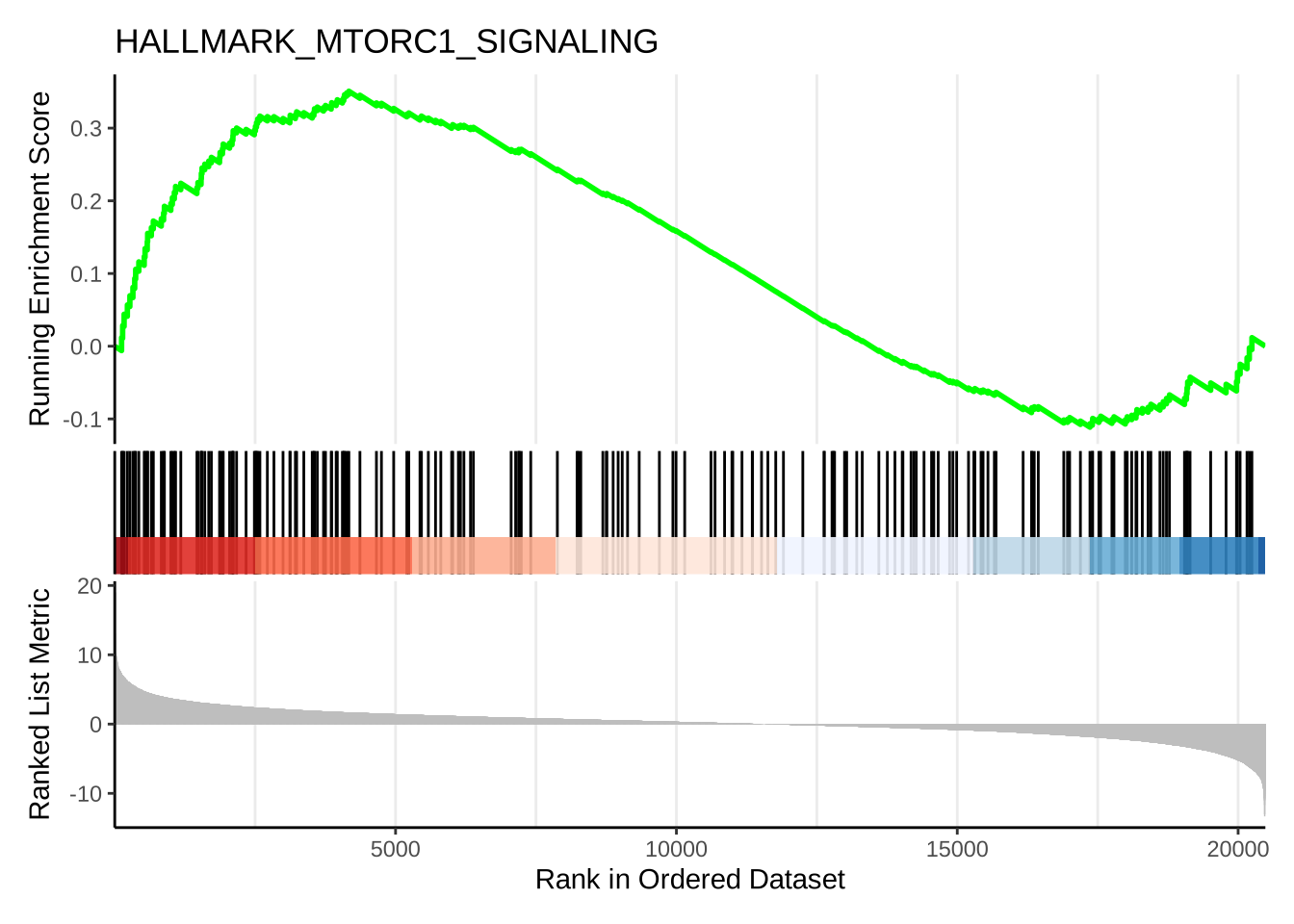
# A barcode plot:
gseaplot2(h_NK_vs_Th,
geneSetID = "HALLMARK_MTORC1_SIGNALING",
title = "HALLMARK_MTORC1_SIGNALING"
)
Extra exercises
# Read in Reactome genes
reactome_gene_sets <- msigdbr(category = "C2", subcategory = "CP:REACTOME")
# Run GSEA with Reactome database
reactome_NK_vs_Th <- GSEA(gl,
minGSSize = 30,
TERM2GENE = reactome_gene_sets[, c("gs_name", "gene_symbol")],
eps = 0, seed = TRUE
)preparing geneSet collections...GSEA analysis...Warning in preparePathwaysAndStats(pathways, stats, minSize, maxSize, gseaParam, : There are ties in the preranked stats (0.07% of the list).
The order of those tied genes will be arbitrary, which may produce unexpected results.leading edge analysis...done...# Count number of significant gene sets
reactome_significant <- length(which(reactome_NK_vs_Th@result$p.adjust < 0.05))
print(paste("Number of significant gene sets with Reactome database is", reactome_significant))[1] "Number of significant gene sets with Reactome database is 53"par(mar = c(5, 20, 3, 3) + 0.1)
# Recode long labels
reactome_NK_vs_Th@result$Description_short <-
reactome_NK_vs_Th@result$Description |>
case_match(
"REACTOME_REGULATION_OF_EXPRESSION_OF_SLITS_AND_ROBOS" ~ "Slits/robos expression regulation",
"REACTOME_RRNA_PROCESSING" ~ "rRNA processing",
"REACTOME_INFLUENZA_INFECTION" ~ "Influenza infection",
"REACTOME_SELENOAMINO_ACID_METABOLISM" ~ "Selenoaminoacid metabolism",
"REACTOME_SRP_DEPENDENT_COTRANSLATIONAL_PROTEIN_TARGETING_TO_MEMBRANE" ~ "SRP-dep. cotranslational membrane targeting",
"REACTOME_RESPONSE_OF_EIF2AK4_GCN2_TO_AMINO_ACID_DEFICIENCY" ~ "EIF2AK4(GCN2) AA deficiency response",
"REACTOME_NONSENSE_MEDIATED_DECAY_NMD" ~ "Nonsense mediated decay NMD",
"REACTOME_EUKARYOTIC_TRANSLATION_INITIATION" ~ "Eukaryotic translation initiation",
"REACTOME_TRANSLATION" ~ "Translation",
"REACTOME_EUKARYOTIC_TRANSLATION_ELONGATION" ~ "Eukaryotic translation elongation"
)
# Bar plot
barplot(rev(-log10(reactome_NK_vs_Th@result$p.adjust[1:10])),
horiz = TRUE, names = rev(reactome_NK_vs_Th@result$Description_short[1:10]),
las = 2, xlab = "-log10(adj.p-value)",
cex.names = 0.7,
col = "lightgreen"
)
abline(v = -log10(0.05))-1.png)
reactome_NK_vs_Th@result$Description_short2<-gsub("_", " ",
gsub("REACTOME_", "",
reactome_NK_vs_Th@result$Description))
reactome_NK_vs_Th_sorted<-reactome_NK_vs_Th@result[order(reactome_NK_vs_Th@result$NES,
decreasing = F),]
reactome_NK_vs_Th_sorted$colors <- ifelse(reactome_NK_vs_Th_sorted$NES > 0, "red", "blue")
par(mar = c(4, 30, 1, 1)) # Make the figure margins larger
barplot(reactome_NK_vs_Th_sorted$NES,
horiz = TRUE, names = reactome_NK_vs_Th_sorted$Description_short2,
las = 2, xlab = "Normalized enrichment score",
cex.names = 0.5,
col = reactome_NK_vs_Th_sorted$colors
)-1.png)
gseaplot(reactome_NK_vs_Th,
geneSetID = "REACTOME_EUKARYOTIC_TRANSLATION_ELONGATION",
title = "Reactome - Eukaryotic translation elongation"
)-1.png)
gseaplot(reactome_NK_vs_Th,
geneSetID = "REACTOME_DAP12_INTERACTIONS",
title = "Reactome - DAP12 interaction"
)-1.png)